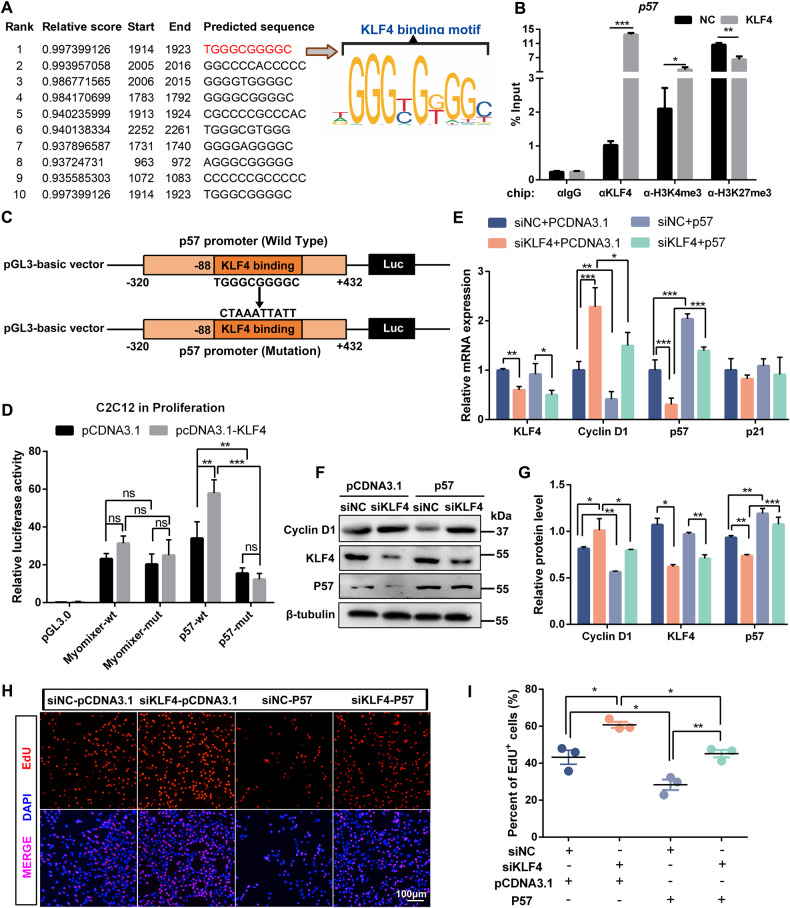
Fig. 6. KLF4 targetedly regulates cell cycle arrest protein P57.
A JASPAR database was used to predict KLF4 binding sites on P57 promoter (−2000bp ~ +300 bp, relative to the TSS of P57 gene). The verified sequence containing KLF4-binding motif in this study was emphasized with red. The KLF4 binding sequence logo created in JASPAR was shown. B ChIP-qPCR analyses of KLF4, H3K4me3, and H3K27me3 enrichment on P57 promoter in control and KLF4 overexpression cells. Data were normalized as a percentage of the input. C Experimental design to amplify the promoter sequence of P57 from −320 bp to +432 bp (relative to the TSS of P57 gene) with the predicted KLF4 binding site (−87bp to −78bp) was mutated, then inserted into the promoter-driven luciferase (Luc) reporter plasmid pGL3 for the dual luciferase assay. D The dual-luciferase reporter assays were performed in C2C12 cells transfected with pCDNA3.1 vector or pCDNA3.1-KLF4 vector using reporter plasmids containing the wild type or mutated P57 promoter (n = 3). E C2C12 cells, cotransfected with si-NC or si-KLF4 and pcDNA3.1 or pcDNA3.1-P57 vector, were cultured in GM for 48 h. The mRNA levels of KLF4, P21, P57, and Cyclin D1 in four groups were detected by qPCR (n = 3). F The protein levels of KLF4, P57, and Cyclin D1 were detected by WB in cells described as (E). G The relative protein levels of target proteins normalized to β-tubulin signals in (F) were obtained through WB band grey scanning. H C2C12 cells were treated as indicated in (E), and EdU staining was performed to compare cell proliferation ability between four experimental groups. Scale bar = 100 μm. I The percentage of EdU-positive cells in (H) was counted in six microscopic fields for each group (n = 3). Data are represented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001 (Student’s t test).