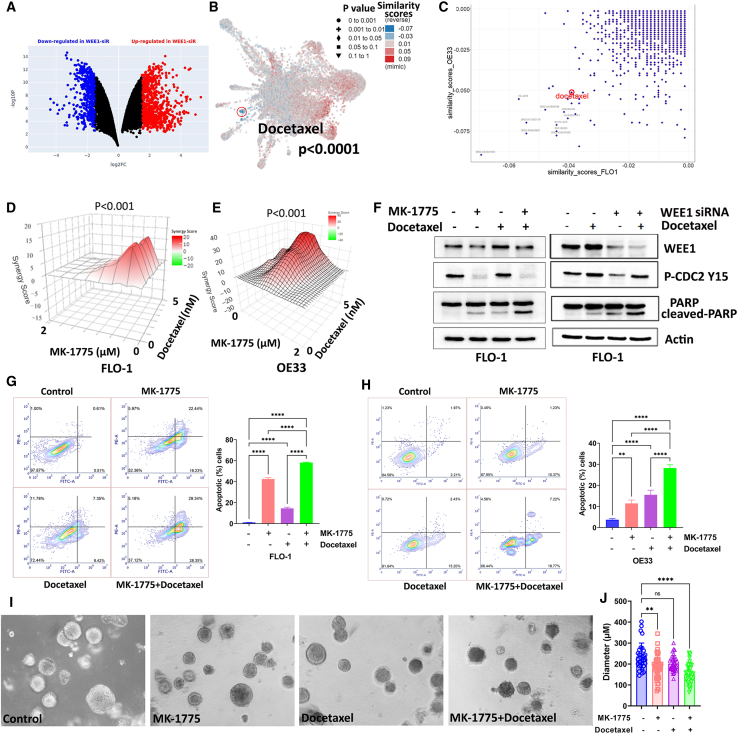
Figure 4.
WEE1 mediates docetaxel resistance in EAC
(A) Differentially expressed genes in OE33 WEE1 siRNA knockdown cells compared with control siRNA transfected cells from RNA sequencing data. Red dots indicate significantly upregulated genes. Blue dots indicate significantly downregulated genes. (B) L1000FWD analysis predicted that docetaxel was one of the drugs with the lowest similarity score to WEE1 siRNA knockdown. (C) Drug similarity scores predicted by L1000FWD based on WEE1 siRNA RNA sequencing data from FLO-1 and OE33 cells. (D and E) SynergyFinder analysis results of FLO-1 and OE33 cells treated with MK-1775 (0–2 μM) and docetaxel (0–5 nM). (F) Left panel: western blot analysis of WEE1, PARP, cleaved PARP, p-CDC2 (Y15), and β-actin in FLO-1 cells treated with MK-1775 1 μM alone, docetaxel 2 nM alone, or a combination. Right panel: similar results in FLO-1 cells treated with docetaxel with or without WEE1 siRNA knockdown. (G and H) Left panels: flow cytometry analysis of Annexin V and PI staining in FLO-1 or OE33 cells treated with MK-1775 1 μM alone, docetaxel 2 nM alone, or a combination. Right panels: quantification of apoptotic cells in the left panels. (I) OE33 spheroids cells treated with MK-1775 alone, docetaxel alone, or a combination. (J) Quantifying data in (F). ∗∗p < 0.01, ∗∗∗∗p < 0.0001.