Figure 3. Relationship between mutational signatures and nucleosome occupancy.
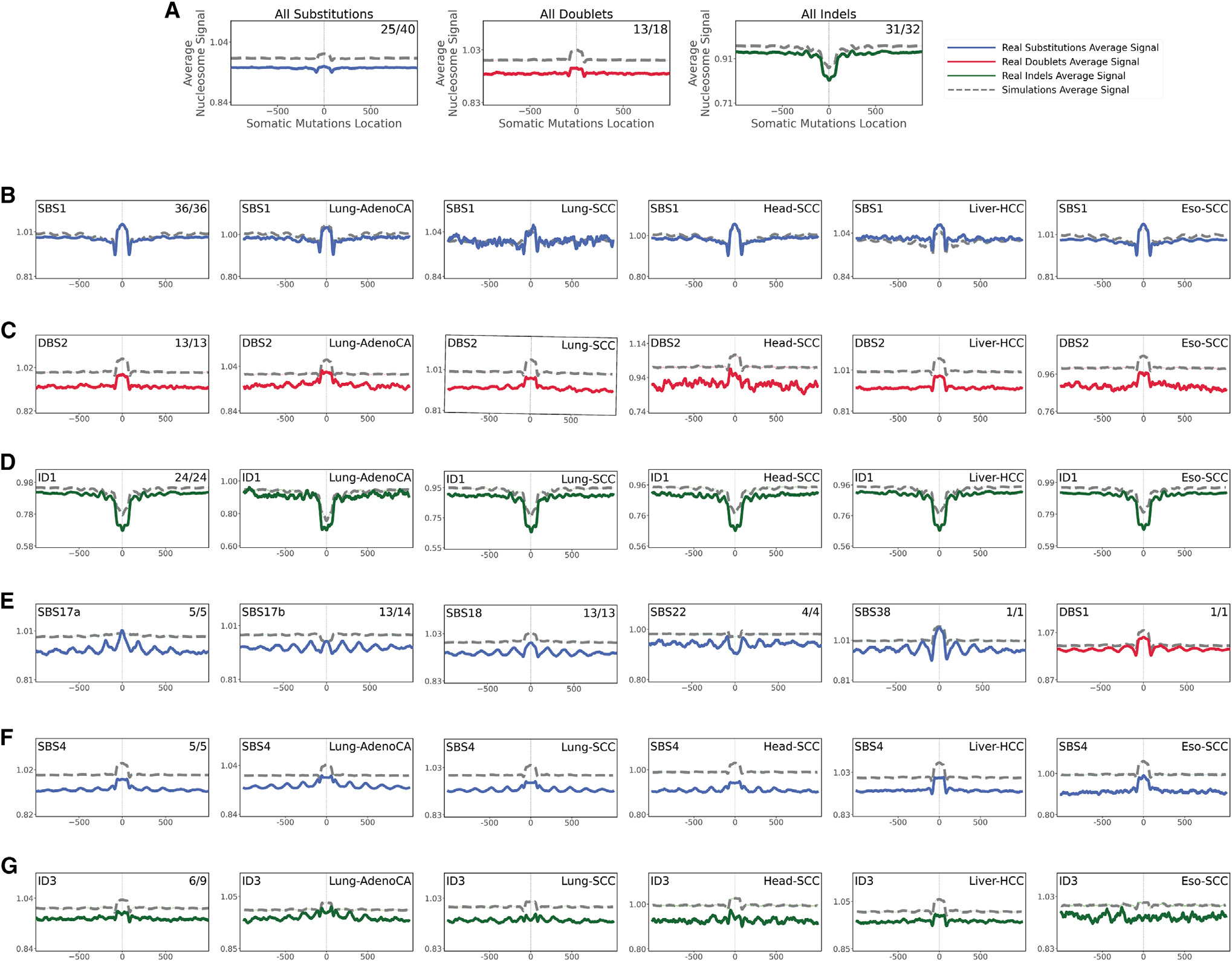
In all cases, solid lines correspond to real somatic mutations, with blue solid lines reflecting SBSs, red solid lines reflecting DBSs, and green solid lines reflecting small IDs. Simulated somatic mutations are shown as dashed lines. Solid lines and dashed lines display the average nucleosome signal (y axes) along a 2 kb window (x axes) centered at the mutation start site for real and simulated mutations, respectively. The mutation site is annotated in the middle of each plot and denoted as 0. The 2 kb window encompasses 1,000 base pairs 5′ adjacent to each mutation as well as 1,000 base pairs 3′ adjacent to each mutation. Where applicable, the total number of similar and considered cancer types using an X/Y format, with X being the number of cancer types where a signature has similar nucleosome behavior (Pearson correlation ≥ 0.5 and adjusted p value ≤ 0.05, z-test corrected for multiple testing using Benjamini-Hochberg) and Y representing the total number of examined cancer types for that signature. For example, signature ID3 in (G) annotated with 6/9 reflects a total of 9 examined cancer types with similar nucleosome behavior observed in 6 cancer types.
(A) All SBSs, DBSs, and IDs across all examined cancer types with each cancer type examined separately.
(B–D) The nucleosome occupancy of signatures SBS1 (B), DBS2 (C), and ID1 (D) are shown across all cancer types as well as within cancers of the lung, head and neck, liver, and esophagus.
(E) Signatures with consistent periodicities of mutation rates around the nucleosome.
(F and G) Tobacco-associated SBS4 (F) and ID3 (G) exhibiting periodicities of mutation rates only in certain cancer types.
See also Figure S3.
