Figure 4. Characterization of DIM interaction with Sub2/DDX39B.
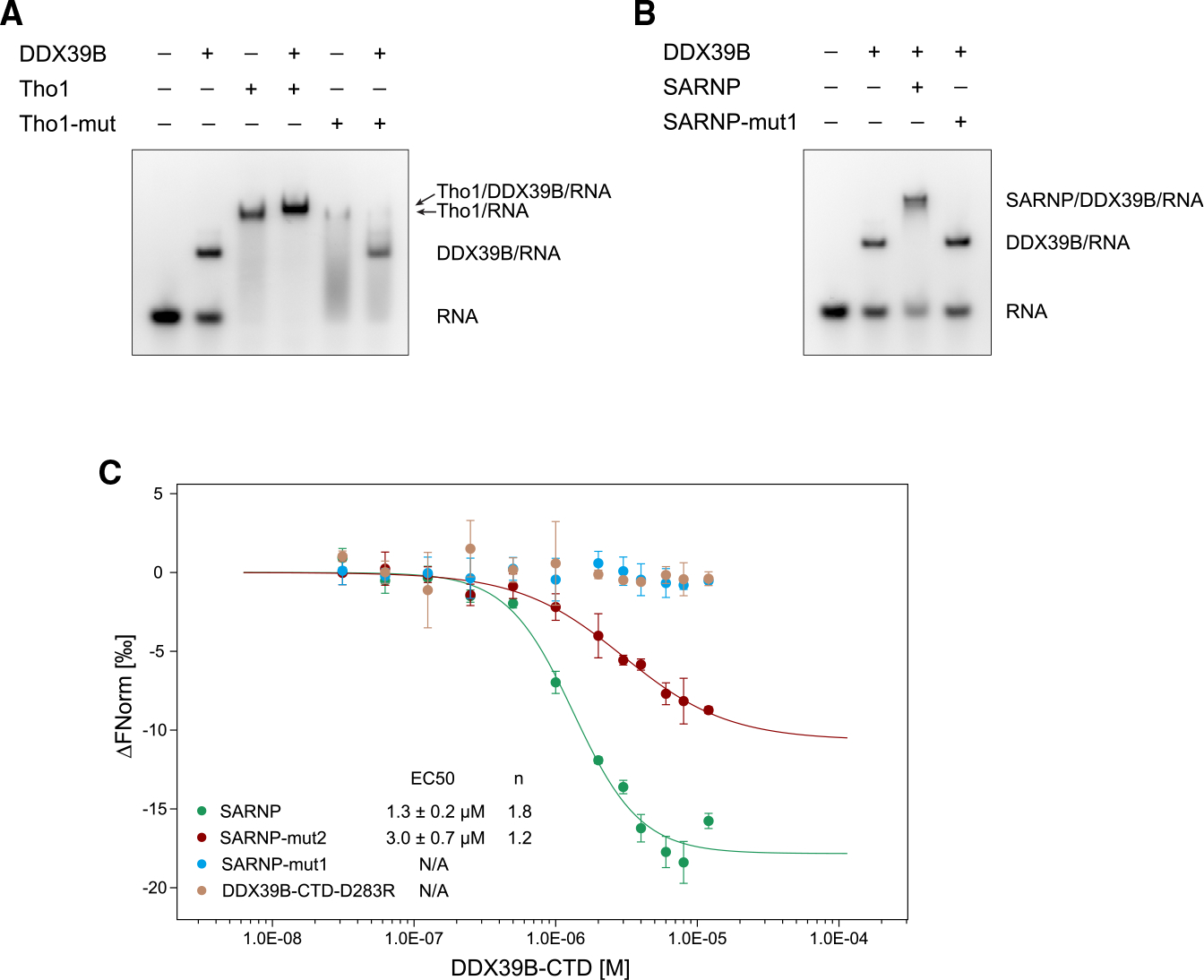
(A) Mutation of the R5 and F9 residues in yeast Tho1 DIMs (Tho1-mut) disrupted binding of DDX39B. EMSA was carried out with poly(U) 15-mer RNA at 100 nM, DDX39B at 0.3 μM, and Tho1 or Tho1-mut (R139A/F143A/R159A/F163A) at 3 μM. Data are representative of three technical replicates.
(B) Mutation of the R5 and F9 residues in all SARNP DIMs (SARNP-mut1) disrupted binding of DDX39B. EMSA was carried out with poly(U) 15-mer RNA at 100 nM, DDX39B at 0.3 μM, and SARNP or SARNP-mut1 (R106A/F110A/R123A/F127A/R153A/F157A/R177A/F181A/R203A/F207A) at 3 μM. Data are representative of three technical replicates.
(C) Microscale thermophoresis (MST) analysis of the SARNP-DDX39B interaction. Measurements of SARNP, SARNP-mut2 (R153A/F157A/R177A/F181A/R203A/F207A, DIM3–5 mutated), or SARNP-mut1 binding to DDX39B-CTD are colored in green, red, and blue, respectively. Measurements of SARNP binding to DDX39B-CTD-D283R are colored in brown. Data were fitted using the Hill equation. EC50 and Hill coefficient (n) are shown. Error bars represent SD from three technical replicates.
