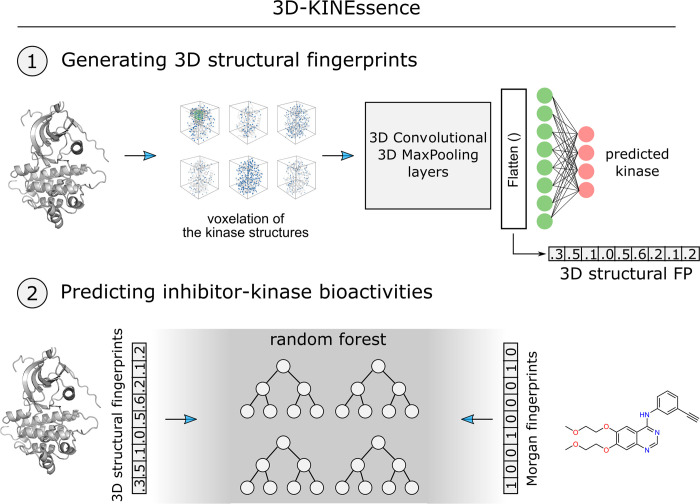
Fig 2. Schematic representation of the 3D-KINEssence pipeline.
The structures of the 69 chosen kinases were obtained from KLIFS [69,70], prepared with Maestro, and gridded with libmolgrid before being provided to 3D convolutional neural networks to train. The features learned in the convolutional layers are flattened and used as 3D convolutional fingerprints (3D FP) in the random forest workflow. The newly generated 3D FP, along with Morgan fingerprints, were used as inputs to the random forest model to predict inhibitor-kinase potency. The representation of the voxels is adapted from Green et al. [85].