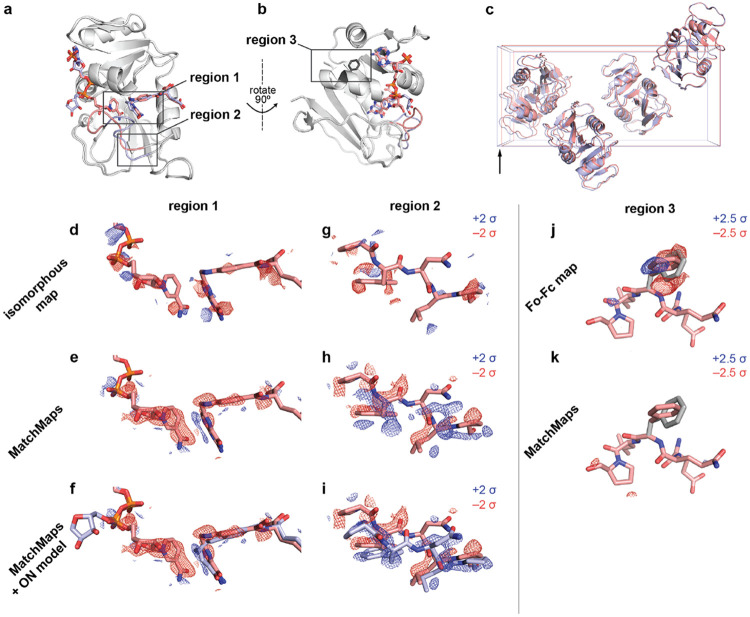
Figure 2: MatchMaps for poorly-isomorphous structures.
(a) The structures 1RX2 and 1RX4 are similar overall (gray cartoons). The structures differ mainly at the active site loop (1RX2, pink cartoon; 1RX4, blue cartoon) and in the positions of the active-site ligands (1RX2, pink sticks; 1RX4, blue sticks). (b) Same as (a), but rotated 90 degrees to the right about the vertical axis (pictured). Sidechain for phenylalanine 103 is shown as dark gray sticks. (c) The unit cells of 1RX2 and 1RX4 differ by 2% along the longest dimension (left to right in this figure) from 98.912 Åto 100.879 Å. (d-f) Visualization of the change in ligand position between 1RX2 (red sticks) and 1RX4 (f, blue sticks). Positive difference density is shown as blue mesh; negative different density is shown as red mesh. Importantly, the 1RX4 structural coordinates were not used in the creation of the the isomorphous or MatchMaps maps. The isomorphous difference map contains essentially no interpretable signal. In contrast, the MatchMaps map (e-f) contains clear signal for disappearance of the cofactor and the lateral sliding of the substrate. (f) is the same as (e), with the addition of the 1RX4 structural coordinates as blue sticks. (g-i) Visualization of the change in loop conformation between 1RX2 and 1RX4. Only protein residues 21-25 are shown. Coloring is as in panels d-f. (g) Again, the isomorphous difference map is not interpretable. (h-i) The MatchMaps positive difference density clearly corresponds with the 1RX4 structural model, which was not used in the creation of the map. (i) is the same as (h), with the addition of the 1RX4 structural coordinates as blue sticks. (j-k) Impact of a spurious conformer on , MatchMaps. 1RX2 model for residues 101-105 are shown as pink sticks. The spurious conformer for Phe103 is shown as gray sticks. (j) map shows clear positive (blue) and negative (red) density recognizing the erroneous conformer as a conformational change. (k) MatchMaps does not show difference density for the spurious conformer.