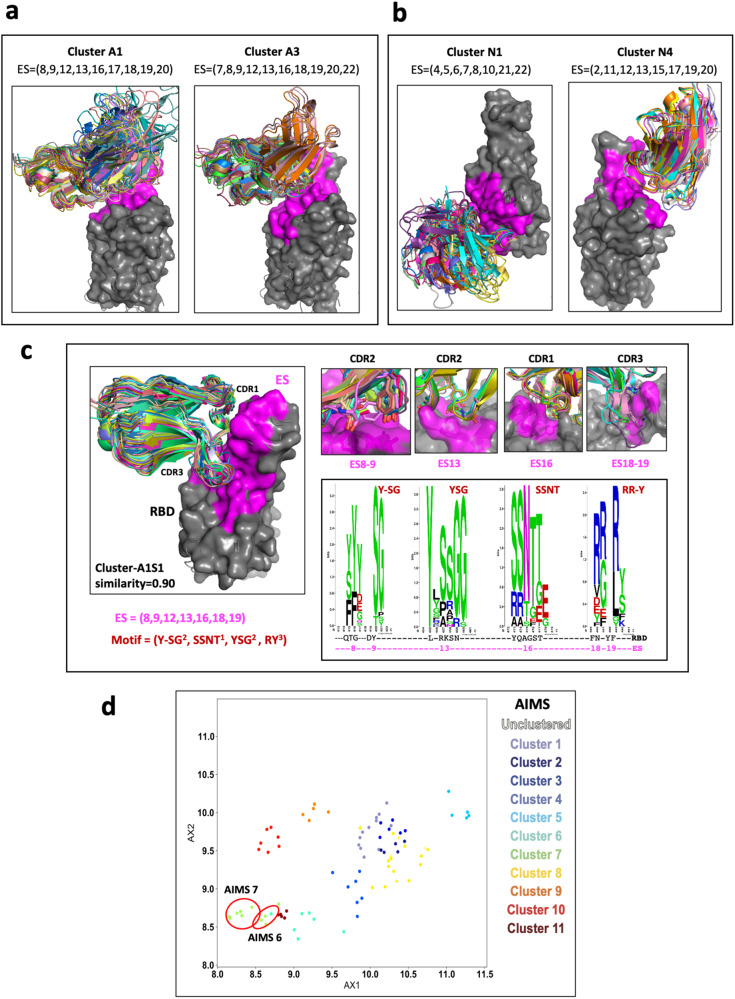
Fig. 5. Identity of the similarity of the ES and clustering of Abs/Nbs (see Supplementary Table 3).
a Illustration of three antibody clusters: A1 and A3, each identifies a specific ES combination. Superimposed are members of the cluster on the RBD (only HV domains are shown for clarity). b Illustration of three nanobody clusters: N1 and N4. RBD is presented as gray surface, magenta indicates the binding areas (footprints) of ES of RBD. c A subset of the Cluster A1, named A1S1, ES = (8,9,13,16,18,19) with similarity ≥ 0.90, shows a strong binding motif on CDR loops. The members (28) of A1S1 are superposed on the RBD on the left panel. On the right top panel are shown the contacts between CDR loops and the binding sites (ES8-9, ES13, ES16, and ES18-19). On the right below panel, WebLogo plots show the amino acids from Abs binding to ES8-9, 13, and ES18-19 respectively. Y,S,G from CDR2 are favor binding to ES13 (RBD residues from 455–460); S,S,N,T of CDR1 favor binding to ES16 (RBD residues 455–459); and R and Y of CDR3 are favor binding to ES18-19 (RBD residues 485-491). d Clustering using AIMS51. Here AX1 and AX2 are “principle components” of biophysical properties, or “mature information”. AIMS6 and AIMS7 are very similar to A1S1.