Abstract
Background
In the first year of life, DNA methylation (DNAm) patterns are established and are particularly susceptible to exposure-induced changes. Some of these changes may leave lasting effects by persistently altering gene expression or cell type composition or function, contributing to disease.
Objectives
In this discovery study, we investigated DNAm associations with sensitization to peanut, egg, or cow’s milk and hypothesized that genes demonstrating DNAm differences in immune cells may play a role in the development of food sensitization.
Methods
Infant sensitization (a skin prick test wheal size that is at least 2 mm greater than the negative control) was measured to peanut, egg, and cow’s milk at age 1 year, and ages of food introduction were reported prospectively. PBMC DNAm was measured in blood samples at 1 year in 144 infants, oversampled for atopy or wheeze. Statistical analysis of Illumina 450k array DNAm data was conducted in R with adjustment for clinical and genetic covariables and a minimum effect size of 1%, false discovery rate of 5%, and medium-confidence false discovery rate threshold of 20%.
Results
There were no DNAm differences between infants with and without peanut, egg, or cow’s milk sensitization. Borderline significant sites with high effect sizes were enriched for methylation quantitative trait loci, hinting at genetic factors influencing DNAm at these sites. DNAm patterns did not differ by peanut or egg introduction before or after 12 months.
Conclusion
This small pilot study did not show differences in methylation by food sensitization or introduction, but it did demonstrate DNAm patterns linked to genetic variants.
Key words: CHILD Cohort Study, DNA methylation, food sensitization, food allergy, epigenetic, peanut, egg, allergenic food, food introduction
Introduction
Previous CHILD Cohort publications support early introduction of potentially allergenic foods for decreasing IgE-mediated food allergy and sensitization in the general population.1,2 Associations between food allergy and dietary introduction imply an environmental or experiential contribution to food allergy development.1,2 Peanut allergy3 and early-life environmental factors4 have also been associated with genome-wide DNA methylation (DNAm) patterns, implying that we may be able to better understand the mechanism underlying the environmental component of food allergy by examining DNAm in children with food allergy or sensitization.
In the first year of life, DNAm patterns are being established and are particularly susceptible to exposure-induced changes. Some of these changes may leave lasting effects by persistently altering gene expression or cell type composition or function, ultimately contributing to disease. In this discovery study, we investigated DNAm associations with sensitization to peanut, egg, and cow’s milk and dietary introduction of peanut and egg at age 1 year. We hypothesized that genes demonstrating DNAm differences in immune cells may play a role in the development of food sensitization.
The CHILD Cohort Study is an observational study of 3455 healthy infants who were recruited from the general population at 4 Canadian sites and enrolled before birth. The REEGLE subset comprises 145 children oversampled for early asthma symptoms.5 Approval was obtained from the University of Manitoba Human Research Ethics Board (HS23871-H2020:192).
Caregivers of CHILD participants prospectively reported their child’s earliest dietary introduction of peanut, egg, and cow’s milk.1 At age 1 year, the children underwent skin prick testing to determine sensitization to peanut, egg, and cow’s milk. A history was taken and physical examination performed for clinical diagnosis of atopic dermatitis during the first year of life.1 Blood was collected and processed to obtain peripheral blood mononuclear cell (PBMC) samples. DNA extraction (using the Qiagen DNAeasy kit, Qiagen, Hilden, Germany), bisulfite conversion (using the Zymo EZ DNAm Gold kit, Irvine, Calif), and DNAm measurement (using the Illumina HumanMethylation450k array, San Diego, Calif) were performed according to the manufacturers' protocols.5 Preprocessing and normalization were accomplished by using the PreprocessNoob and BMIQ functions5; 424,644 probes remained after filtering (based on a detection P value of >.01 and/or bead count of <3, probes with known single-nucleotide polymorphisms, probes localized to sex chromosomes, and cross-reactive probes).5
Estimations of blood cell type proportions were generated by using the FlowSorted.CordBloodCombined.450k6 reference set using the estimateCellCounts2 function with the default cord blood probe selection parameter "any" to estimate the relative proportions of 5 cell types (CD4 T cells, CD8 T cells, B cells, natural killer cells, and monocytes) in PBMC samples collected at age 1 year. CLR-transformation followed by principal component analysis of estimated PBMC proportions was performed to generate principal components, and the first 4 principal components were used in linear regressions to account for cell type differences. The 565 unique probes used to estimate blood cell proportions were removed, resulting in 424,079 remaining probes. Surrogate variables were used to capture variance due to batch effects, along with variance caused by unknown biologic variables. Surrogate variables were created by using the sva package; we included 18 surrogate variables based on recommendation from the estdim function.
Using the limma R package,7 we performed 3 separate epigenome-wide analyses of peripheral blood to evaluate DNAm associations with sensitization to peanut, egg, or cow’s milk at age 1 year, and peanut and egg introduction, respectively, by age 1 year.
Each model was adjusted for covariates previously associated with DNAm or food sensitization or allergy, including sex at birth, self-reported race and ethnicity, and cell type proportions. We included 18 surrogate variables for each model to capture batch effects and unmeasured biologic variation. Changes in DNAm were considered significant if they displayed an absolute effect size (difference in DNAm β-value between groups) greater than 1% and surpassed a 5% false discovery rate (FDR).7, 8, 9 To explore other potential sites of differential DNAm, we also included a "medium-confidence" 20% FDR threshold.
Results and discussion
Of the infants in the study, 58% were male and 42% were female at birth (see Table I for further demographic characteristics). Of the 124 infants with complete data, 27 (22%) had a positive skin prick test result to peanut, egg, or cow’s milk.
Table I.
Participant demographics of the CHILD REEGLE subset
| Characteristic | Sensitization to peanut, egg, or cow's milk at age 12 mo (n = 124) | Peanut dietary introduction by age 12 mo (n = 123) | Egg dietary introduction by age 12 mo (n = 129) |
|---|---|---|---|
| Characteristic, no. (%) | |||
| Yes | 27 (22) | 52 (42) | 98 (76) |
| No | 97 (78) | 71 (58) | 31 (24) |
| Sex at birth, no. (%) | |||
| Female | 52 (42) | 53 (43) | 56 (43) |
| Male | 72 (58) | 70 (57) | 73 (57) |
| Maternal self-reported race and ethnicity, no. (%) | |||
| Asian | 15 (12) | 12 (10) | 14 (11) |
| Black/African American | 0 (0) | 0 (0) | 0 (0) |
| Hispanic | 2 (1.5) | 2 (1.5) | 2 (1.5) |
| Indigenous | 7 (5.5) | 7 (5.5) | 7 (5) |
| Middle Eastern | 2 (1.5) | 2 (1.5) | 2 (1.5) |
| Other | 3 (2.5) | 3 (2.5) | 4 (3) |
| White | 95 (77) | 97 (79) | 100 (78) |
| Paternal self-reported race and ethnicity, no. (%) | |||
| Asian | 12 (10) | 10 (8) | 12 (9) |
| Black/African-American | 0 (0) | 0 (0) | 0 (0) |
| Hispanic | 1 (1) | 1 (1) | 1 (0.8) |
| Indigenous | 2 (1.5) | 1 (1) | 1 (0.8) |
| Middle Eastern | 2 (1.5) | 2 (2) | 2 (1.6) |
| Other | 9 (7) | 10 (8) | 10 (7.8) |
| White | 98 (79) | 99 (80) | 103 (80) |
| Moderate-to-severe eczema, no. (%) | |||
| Yes | 8 (6) | 6 (5) | 7 (5) |
| No | 116 (94) | 117 (95) | 122 (95) |
| Breast-feeding duration no. (%) | |||
| Did not breast-feed | 7 (6) | 10 (8) | 13 (10) |
| 0.5-6 mo | 29 (23) | 27 (22) | 28 (22) |
| 7-12 mo | 43 (35) | 42 (34) | 43 (33) |
| 13-24 mo | 45 (36) | 44 (36) | 45 (35) |
| Maternal stress self-reported at 6 mo (no.), mean ± SD∗ | 13 ± 7 | 13 ± 7 | 13 ± 7 |
| Predicted cell type proportions, mean ± SD | |||
| CD4+ T lymphocytes | 0.431 ± 0.0778 | 0.431 ± 0.0778 | 0.431 ± 0.0778 |
| Natural killer cells | 0.0802 ± 0.0365 | 0.0802 ± 0.0365 | 0.0802 ± 0.0365 |
| B lymphocytes | 0.192 ± 0.0686 | 0.192 ± 0.0686 | 0.192 ± 0.0686 |
| Monocytes | 0.166 ± 0.0699 | 0.166 ± 0.0699 | 0.166 ± 0.0699 |
| CD8+ T lymphocytes | 0.156 ± 0.0478 | 0.156 ± 0.0478 | 0.156 ± 0.0478 |
The maternal stress scale comprises scores from the Perceived Stress Scale. Individual scores are integers ranging from 0 to 40. Higher scores indicate that mothers reported higher perceived stress.
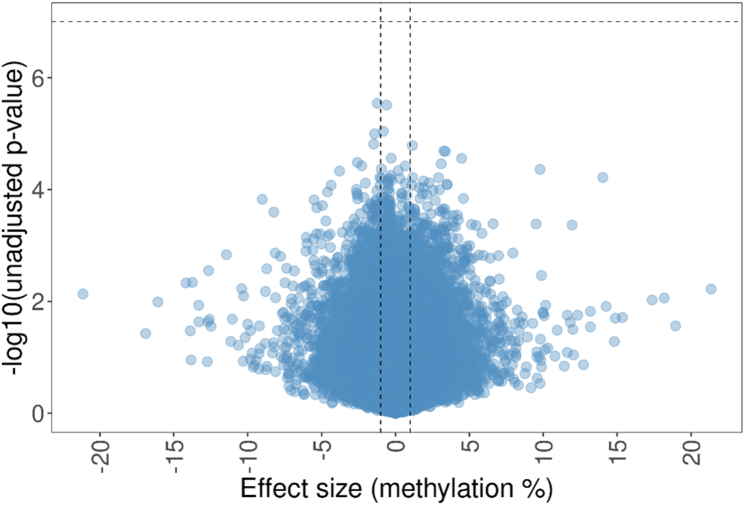
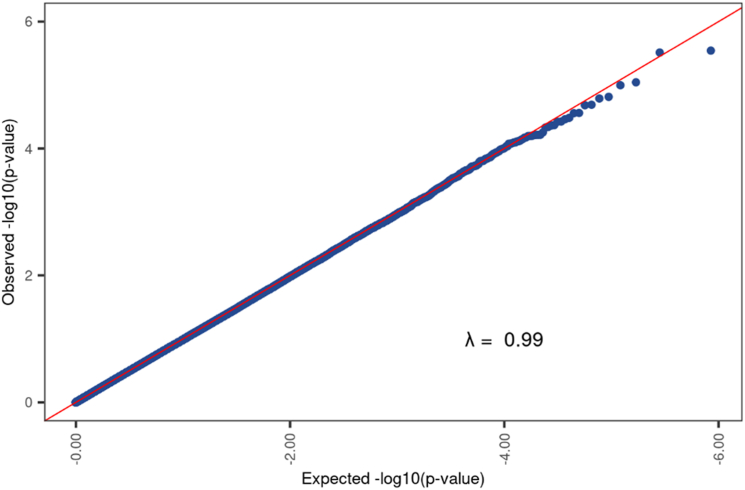
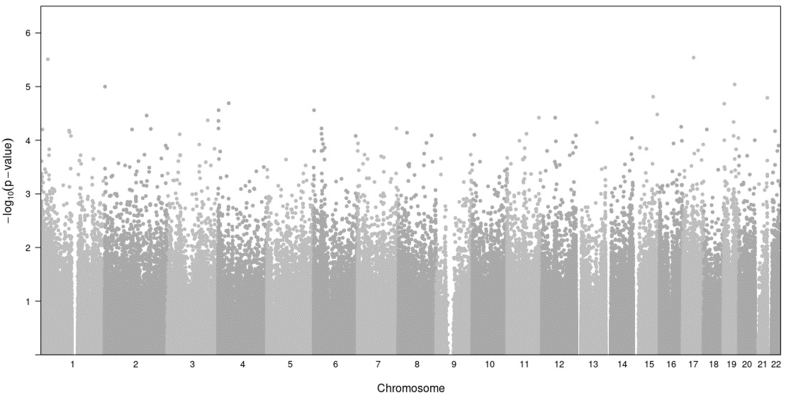
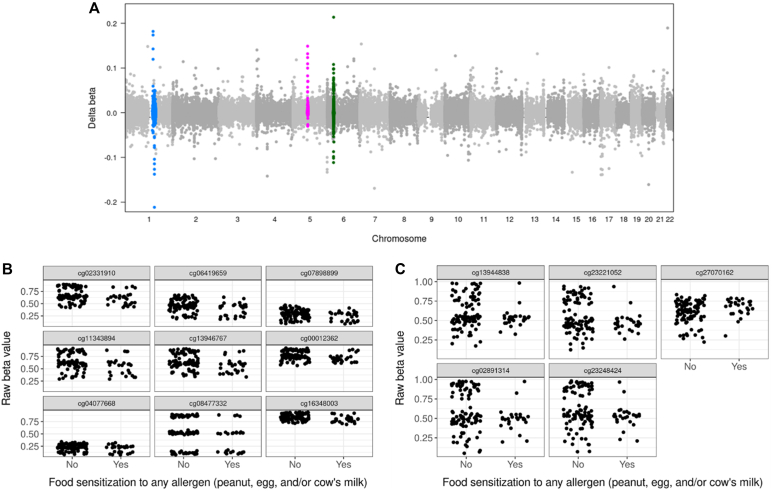
We found no differences in peripheral blood DNAm between participants with and without sensitization to peanut, egg, or cow’s milk at 5% or 20% FDR (Fig 1 and 2). On inspection of Manhattan plots, we noted unusual patterns of tightly clustered small P values on several chromosomes (Figs 3 and 4, A). Closer examination of these groups revealed a high prevalence of trimodally distributed β-values (Figs 4, B and C), indicating that DNAm at these sites may be influenced by nearby genetic variants (see Table E1 in the Online Repository at www.jaci-global.org).
Fig 1.
No significant differences were found in DNAm between infants with and without sensitization to peanut, egg, or cow’s milk. Vertical lines display the 1% methylation effect size threshold. Horizontal lines display the 20% FDR threshold.
Fig 2.
The model comparing infants with and without sensitization to peanut, egg, or cow’s milk has values falling within the expected range. The λ statistic identifies whether points in the QQplot are within the expected range (λ = 1) or whether the P values are greater, or more significant, than expected (λ > 1). In our case, a λ value of 0.99 points to a good linear fit for the model with values falling within the expected range. Model denoted as (∼0 + food_sens_any + sex_at_birth + maternal_race_ethnic + paternal_race_ethnic + stress + breast-feeding duration + cell type + 18 surrogate variables).
Fig 3.
Analysis of CpG sites across the genome in comparison with their DNAm change significance. The Manhattan plot shows the locations of CpGs within the genome and their associated significance value determined through the difference between infants with and without sensitization to peanut, egg, or cow’s milk. None of the CpG sites met the FDR thresholds of 5% or 20%.
Fig 4.
Clusters of CpGs with high effect sizes for food sensitization association despite non–statistically significant test results may be suggestive of possible genetic effects. A, Clustered CpG sites (examples are highlighted with color) signify DNA sites in close proximity to one another with frequent high effect sizes in the food sensitization analysis. Chromosome 1 (B) and chromosome 5 (C) CpG cluster regions show trimodal distribution of DNAm, likely owing to nearby genetic variants influencing methylation levels.
Among the infants with complete dietary introduction data, 52 of 123 (42%) and 98 of 129 (76%) had reported peanut and egg introduction before age 12 months, respectively (Table I). At FDRs of 5% and 20%, we observed no differences in DNAm between participants who introduced peanut (see Fig E1, A in the Online Repository at www.jaci-global.org) or egg (Fig E1, B) before versus after age 12 months. We did not observe any clustering of small P values in Manhattan plots of early peanut or egg introduction (see Fig E2 in the Online Repository at www.jaci-global.org).
This study is one of the largest to examine DNAm differences between infants with and without sensitization to peanut, egg, or cow’s milk, and with timing of dietary introduction of potentially allergenic foods. DNAm differences were not significantly associated with food sensitization. However, we did note an enrichment of potentially genetically controlled sites of DNAm with high effect sizes, which supports a polygenic model of food sensitization risk and is consistent with the findings of previous research.
One previous study examined 125 targeted genomic regions associated with food sensitization and found that 12 of them were genetically influenced in twins with (n = 10) and without (n = 10) peanut allergy.3 In a study examining participants with allergy (n = 44) and age-matched control participants without allergy (n = 21), DNAm profiles at a specific gene locus were influenced by genotype; however, loss of methylation at the gene associated with food allergy was not substantially influenced by genetic variation within the single-nucleotide polymorphisms tested.8 These studies3,7,8 have shown that the alteration of gene expression caused by DNAm changes can result in modified PBMC function, such as in TH1/TH2 lymphocytes.
One of our study’s strengths was the prospectively collected cohort data allowing an unbiased approach to DNAm changes in infants with or without sensitization at age 1 year. The evaluation of sensitization and dietary introduction of highly allergenic foods and DNAm at age 1 year was ideal timing because DNAm patterns are being established and are particularly susceptible to exposure-induced changes in the first year of life.
The limitations of this study included the moderate sample size for a DNAm study. The relatively low number of participants displaying sensitization to each allergen required grouping of infants with food sensitization to peanut, egg, or cow’s milk to assess general epigenetic associations with food sensitization rather than sensitization to each specific food allergen. We did not have the power to evaluate potential epigenetic associations with clinically manifested IgE-mediated food allergy, which may have greater biologic plausibility than associations with sensitization.
In the general population CHILD Cohort, highly allergenic food sensitization and delayed introduction1,2 were not associated with DNAm. Other evidence has suggested epigenetic modifications are a mechanism by which exposures in childhood may be linked to molecular events that cause disease,3,8 and our study may have been underpowered to detect a gene-environment interaction with food sensitization and dietary introduction. DNAm patterns of infants with and without sensitization to peanut, egg, or cow’s milk at age 1 year demonstrated linkages to genetic variants. With further analysis, we can determine whether DNAm levels at certain CpG sites are associated with genetic variants that contribute to a higher or lower likelihood of developing food sensitization. Our study was discovery based and should be considered hypothesis generating. Further studies are needed to identify areas of the genome that may contribute to food allergy and sensitization either through genetic variants or epigenetic modifications.
Disclosure statement
Supported by funding from the Andison Family Foundation, as well as by core funding for the CHILD Cohort Study from The Allergy, Genes and Environment Network of Centres of Excellence (AllerGen NCE) and the Canadian Institutes of Health Research (CIHR).
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Acknowledgments
We thank the CHILD Cohort Study participants and their families and the CHILD Cohort Study team members at each site; the dedication of these participants and team members made this study possible.
Supplementary data
References
- 1.Simons E., Balshaw R., Lefebvre D.L., Dai D., Turvey S.E., Moraes T.J., et al. Timing of introduction, sensitization, and allergy to highly allergenic foods at age 3 years in a general-population Canadian cohort. J Allergy Clin Immunol Pract. 2020;8:166–175.e10. doi: 10.1016/j.jaip.2019.09.039. [DOI] [PubMed] [Google Scholar]
- 2.Azad M.B., Dharma C., Simons E., Tran M., Reyna M.E., Dai R., et al. Reduced peanut sensitization with maternal peanut consumption and early peanut introduction while breastfeeding. J Dev Orig Health Dis. 2021;12:811–818. doi: 10.1017/S2040174420001129. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X., Han X., Lyu S.-C., Bunning B., Kost L., Chang I., et al. Targeted DNA methylation profiling reveals epigenetic signatures in peanut allergy. JCI Insight. 2021;6 doi: 10.1172/jci.insight.143058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Z., Liu Y. DNA methylation in human diseases. Genes Dis. 2018;5:1–8. doi: 10.1016/j.gendis.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sbihi H., Jones M.J., MacIsaac J.L., Brauer M., Allen R.W., Sears M.R., et al. Prenatal exposure to traffic-related air pollution, the gestational epigenetic clock, and risk of early-life allergic sensitization. J Allergy Clin Immunol. 2019;144:1729–1731.e5. doi: 10.1016/j.jaci.2019.07.047. [DOI] [PubMed] [Google Scholar]
- 6.Salas L., Gervin K., Jones M. FlowSorted.CordBloodCombined.450k: Illumina 450k/EPIC data on FACS and MACS umbilical blood cells. 2019. Bioconductor; Open Source Software for Bioinformatics. https://bioconductor.org/packages/release/data/experiment/html/FlowSorted.CordBloodCombined.450k.html Available at:
- 7.Martino D., Joo J.E., Sexton-Oates A., Dang T., Allen K., Saffery R., et al. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy. Epigenetics. 2014;9:998–1006. doi: 10.4161/epi.28945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martino D., Neeland M., Dang T., Cobb J., Ellis J., Barnett A., et al. Epigenetic dysregulation of naive CD4+ T-cell activation genes in childhood food allergy. Nat Commun. 2018;9:3308. doi: 10.1038/s41467-018-05608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dijk F.N., Xu C., Melén E., Carsin A.-E., Kumar A., Nolte I.M., et al. Genetic regulation of IL1RL1 methylation and IL1RL1-a protein levels in asthma. Eur Respir J. 2018;51:1701377. doi: 10.1183/13993003.01377-2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.