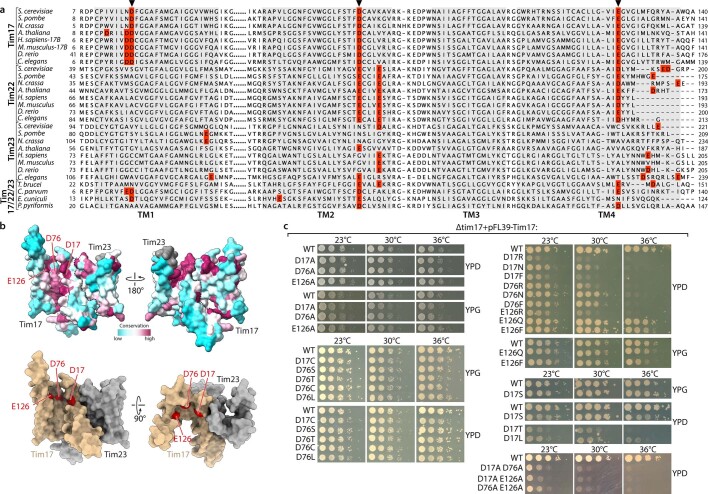
Extended Data Fig. 5. Sequence alignment of Tim17 protein family with conserved negative charges within the TMDs and growth phenotype of corresponding tim17 mutants.
a, Sequence alignment of Tim17 from Saccharomyces cerevisiae, Schizosaccharomyces pombe, Neurospora crassa, Caenorhabditis elegans, Drosophila melongaster, Danio rerio, Homo sapiens, Mus musculus and Arabidopsis thaliana (paralog 1) generated by TMcoffee60. Predicted TMDs were assigned following sequence alignment of S. c. Tim17 with S. c. Tim22 and according to the cyro-EM structure of Tim22 (6LO8)25. Grey boxes, predicted transmembrane (TM) domains; Red, highly conserved negatively charged residues within predicted TM domains; Arrowheads, negatively charged residues analysed in this study. b, Tim17 and Tim23 amino acid sequence conservation mapped onto the surface of the ColabFold predicted Tim17-Tim23 heterodimer (upper panel). Space filling model of the Tim17-Tim23 heterodimer with the conserved negatively charged acidic patch formed by D17, D76 and E126 indicated (lower panel). c, Growth analysis of WT and tim17 mutants in tim17Δ background related to Extended Data Table 1. Yeast strains expressing WT or the indicated tim17 mutant variant were grown on non-fermentable (glycerol, YPG) or fermentable (dextrose/glucose, YPD) agar medium at the indicated temperatures.