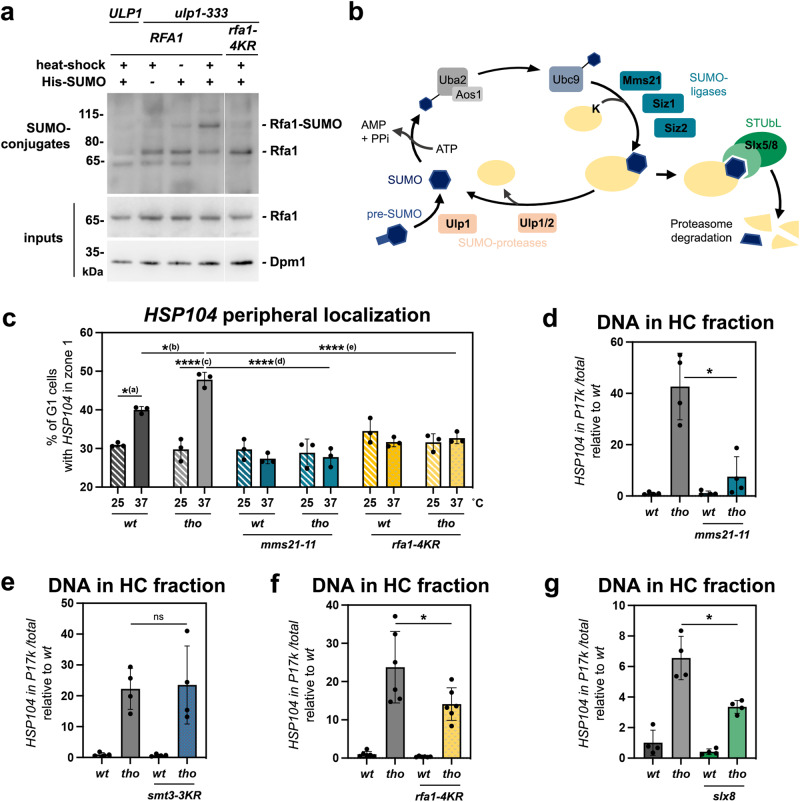
Fig. 4. The SUMOylation pathway is involved in R-loop-dependent repositioning to NPCs.
a Western blot detection of Rfa1 in input fractions (bottom panel) or purified SUMO-conjugates (top panel) obtained from the indicated strains. Cells carrying the His-SMT3 (His-SUMO) construct as indicated were grown at 25 °C or heat shocked at 37 °C for 15 min (heat shock). The positions of Rfa1 species are indicated, as well as molecular weights (kDa, kilodaltons). The apparent molecular mass of the Rfa1-SUMO species is ~90 kDa, which is consistent with mono-SUMOylation (Rfa1: 70 kDa; apparent molecular weight of SUMO: 15–20 kDa). One representative experiment (out of three) is displayed; the two other biological replicates are featured in Supplementary Fig. 4a, b. Dpm1 is used as a loading control for input fractions. b Overview of the components of the SUMO pathway in S. cerevisiae. c Fraction of G1 cells (%) showing HSP104 in zone 1 (mean ± SD, n = 3 independent experiments) in the indicated strains grown at 25 °C or heat shocked at 37 °C for 15 min. Statistical test: two-sided Fisher’s exact test; P-values were calculated on the total number of counted cells (between 287 and 409 cells/condition); (a), p = 1.59 × 10−2; (b), p = 2.45 × 10−2; (c), p = 4.96 × 10−7; (d), p = 2.84 × 10−8; (e), p = 4.43 × 10−5. Values for wt and tho mutant cells (in gray) are the same as in Fig. 3h. d–g qPCR-based quantification of the amount of DNA from the HSP104 locus in heavy chromatin (HC) fractions from the indicated strains heat shocked at 37 °C for 15 min (% of HSP104 in P17K relative to total [S17K + P17K]; mean ± SD, n = 4 independent experiments for panels d, e, g, n = 6 independent experiments for panel f, relative to wt). Statistical test: two-sided Mann-Whitney-Wilcoxon test; d, *p = 2.86 × 10−2; f, **p = 4.11 × 10−2; g, *p = 2.86 × 10−2. Note that experiments involving mms21-11, smt3-3KR or rfa1-4KR mutants are performed with isogenic W303 derivatives, in which the co-fractionation phenotype is reproducibly more pronounced than in other genetic backgrounds. Source data are provided as a Source Data file.