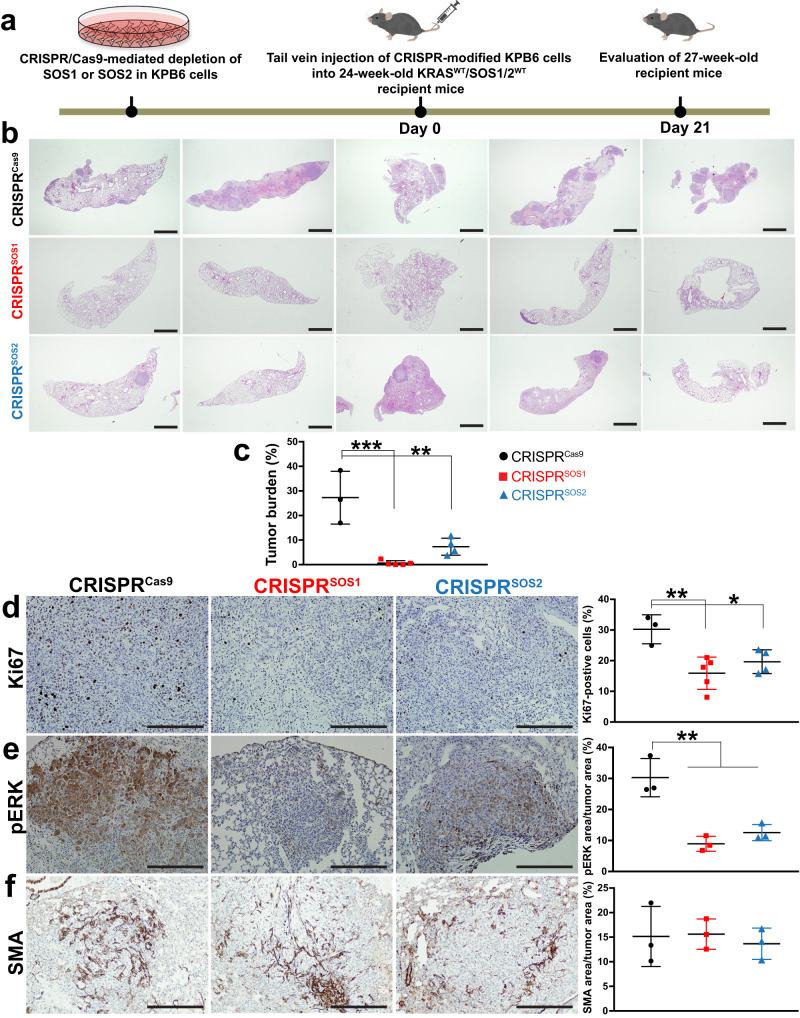
Fig. 6. Nesting and progression of KPB6 lung tumor cells devoid of SOS1 or SOS2 in the lungs of WT syngeneic mice.
a Schematic illustration of experimental regime and timing of tail vein injection of CRISPR/Cas9-modified KPB6 cells devoid of SOS1 or SOS2 into wild-type KRASWT/SOS1/2WT mice. b Representative images of H&E-stained, paraffin-embedded sections from the 5 different lobes of lungs of 27-week-old, SOS1/2WT/KRASWT mice at 3 weeks after receiving intravenous injection of 1 × 105 CRISPRCas9, CRISPRSOS1 and CRISPRSOS2 KPB6 cells. c Bar graph showing the relative percentage of lung tumor burden in each experimental group. d Representative images of paraffin-embedded sections from lung tumors of 27-week-old wild-type mice SOS1/2WT/KRASWT that were injected with CRISPRCas9, CRISPRSOS1 and CRISPRSOS2 cells, after immunolabeling for Ki67. d–f The graphs on the right side show the relative percentage Ki67-positive cells (d), pERK levels (e), and SMA-immunopositive CAFs (f) in the tumors detected in each of the indicates genotypes and experimental conditions in the three experimental groups. Data shown as mean ± SD. In (c): ***P = 0.0002 and **P = 0.0026. In (d): **P = 0.0062 and *P = 0.040. In (e): **P = 0.0017 vs SOS1KO and **P = 0.0045 vs SOS2KO. One-way ANOVA and Tukey’s test. c, d n = 3 independent mice for CRISPRCas9, n = 5 independent mice for CRISPRSOS1 groups and n = 4 independent mice for CRISPRSOS2 group. e, f n = 3 independent mice for each genotype. Scale bars: b 1 mm and d–f 100 μm. Source data are provided as a Source data file.