Figure 7: Activation of JNK signaling in the background of M1BP downregulation promotes autophagy.
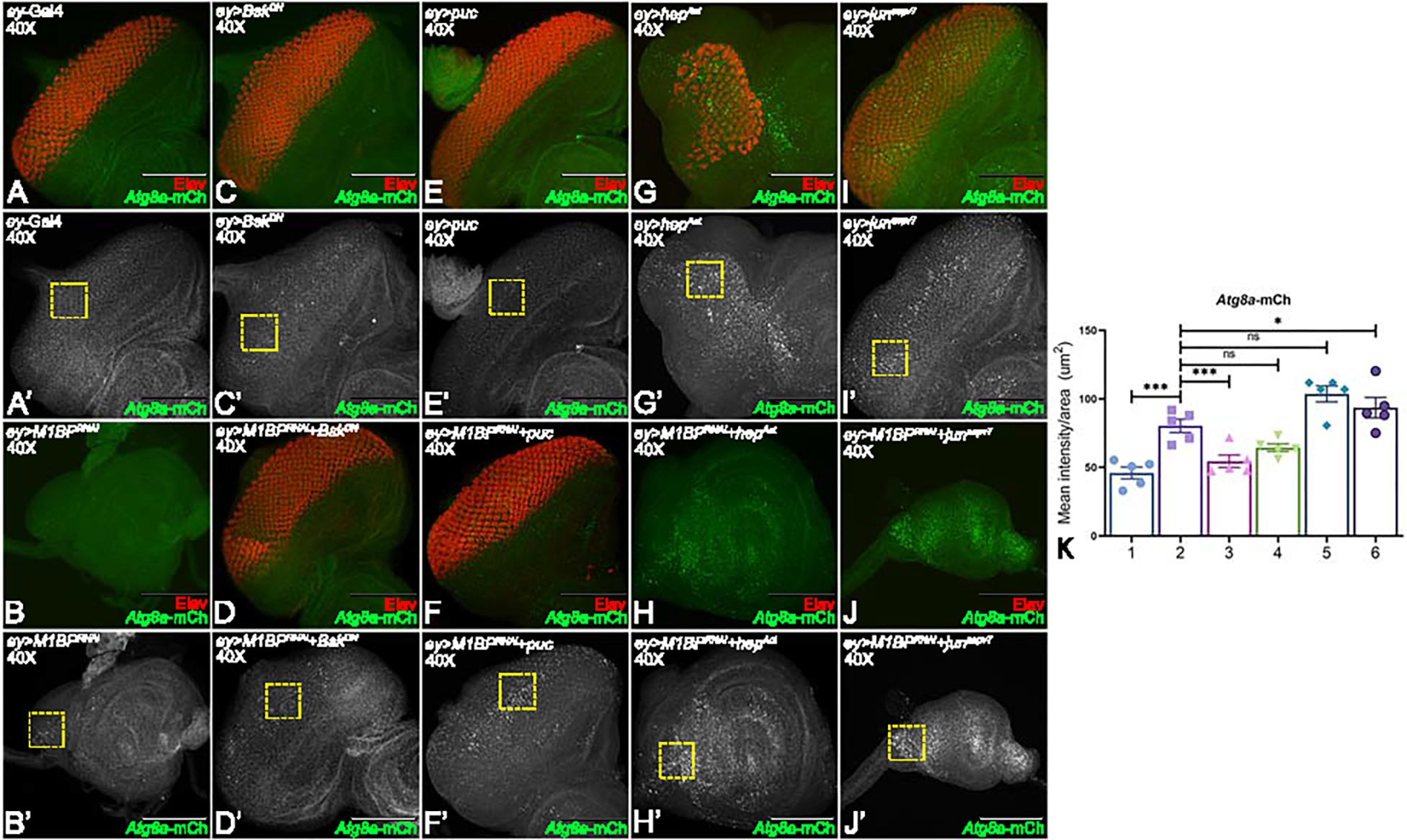
(A-J) Eye-antennal imaginal discs of (A, A’) ey-Gal4 control, (B, B’) ey>M1BPRNAi, (C, C’) ey>bskDN, (D, D’) ey>M1BPRNAi+bskDN, (E, E’) ey>puc, (F, F’) ey>M1BPRNAi+puc, (G, G’) ey>hepAct, (H, H’) ey>M1BPRNAi+hepAct, (I, I’) ey>junaspv7, (J, J’) ey>M1BPRNAi+junaspv7 were assessed for changes in Atg8a-mCherry (green) and pan neuronal marker Elav (red) expression. (A’-J’) Eye antennal imaginal disc showing split channel for Atg8a-mCherry expression. (C-F) JNK pathway was downregulated using bskDN (C, C’) ey>bskDN and puc (E, E’) ey>puc. (G-J) JNK pathway was activated using hepAct (G, G’) ey>hepAct and junaspv7 (I, I’) ey>junaspv7. (K) Quantification of Atg8a-mCherry intensity using Fiji/ImageJ software (NIH). Quantification was performed using standard 100X100 pixels ROI. ROI is shown as yellow dashed boxes. Graph was plotted with mean +/− SEM. The genotypes depicted in the graph are 1: ey-Gal4, 2: ey>M1BPRNAi, 3: ey>M1BPRNAi+bskDN, 4: ey>M1BPRNAi+puc, 5: ey>M1BPRNAi+hepAct and 6: ey>M1BPRNAi+junaspv7. Statistical significance in each graph is shown by p-value: ****p<0.0001, ***p<0.001; **p<0.01; *p<0.05. The orientation of all imaginal discs is identical with posterior to the left and dorsal up. The magnification of all eye-antennal imaginal discs is 40X.
