Figure 2.
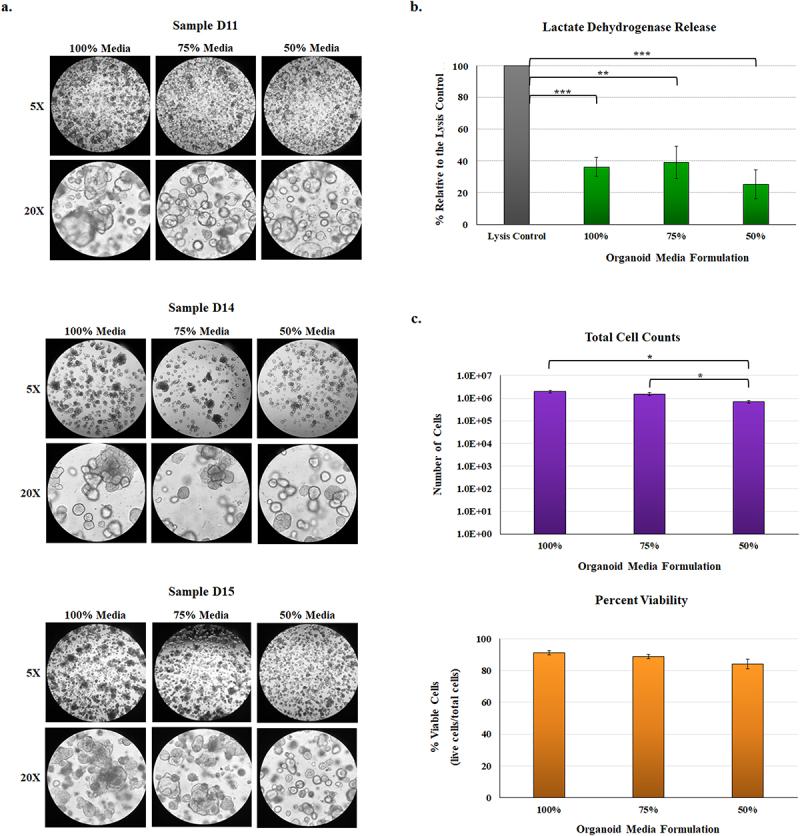
Phenotypic analysis of malnourished organoids.
a. Phenotypic evaluation. Following culturing in the three media formulations, organoids derived from three separate donor samples (D11, D14, and D15) were evaluated phenotypically for changes in appearance. Overall and despite the reduced nutrient concentrations in the malnourished media, organoids matured and developed the characteristic sphere shape. Images above were examined at 5X (top) and 20X (bottom) magnifications for all three sets of samples. The images represent three independent experiments, each with two technical duplicates, for the three organoid lines. The data indicate that organoid development in malnourished conditions is consistent across the three separate donor samples.
b. Lactate dehydrogenase (LDH) release. Culture supernatants were collected from D11, D14, and D15 organoids following seven days of culture in the 100%, 75%, or 50% media formulations. The data represent the average of four biologically independent experiments from all three lines ± the standard errors of the mean (SEM), and are plotted relative to the positive lysis control provided by the kit. All samples had significantly less LDH release relative to the positive control (**, p <.01; ***, p <.001). There were no significant differences in LDH release across the organoid media formulations.
c. To examine cellular viability, D14 organoids were cultured in the three media formulations for seven days. Subsequently, the organoids were trypsinized and stained with trypan blue to obtain total cell numbers and the percent viability of the cells, calculated as the percent of live cells relative to the total number of cells. The 50% media formulation had reduced total cell counts (*, p <.05) compared to the 100% and 75% media formulations. However, there were no significant differences in the percent viability across the treatments. For both analyses, data represent the average of three biologically independent experiments ± the SEM, and each experiment had three technical replicates.
