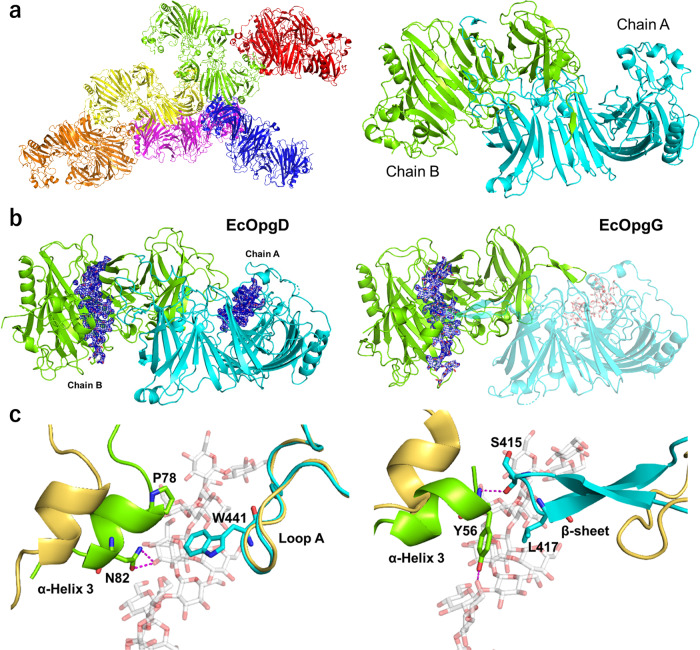
Fig. 2. Structures of EcOpgD and EcOpgG.
a Overall structure of ligand-free EcOpgD. (left) Structure of an asymmetric unit. There are 12 monomers in an asymmetric unit with almost identical conformations (RMSD within 0.30 Å). The free energy of assembly dissociation (ΔGdiss) was calculated by PISA32. All ΔGdiss values of dimer interfaces in the asymmetric unit are higher than 32.9 kcal/mol, indicating that the dimer is a stable assembly. Thus, the six dimers are shown in red, blue, magenta, light green, yellow, and orange. (right) Biological assembly. Chains A and B are shown in cyan and light green, respectively. The RMSD between EcOpgD and EcOpgG (PDB: 1txk) is 2.206 Å. b Overall structures of Michaelis complexes of the EcOpgD D388N (left) and EcOpgG D361N (right) mutants. Chains A and B are shown in cyan and light green, respectively. The two subunits represent a biological assembly. The electron densities of β-1,2-glucans are shown as Fo–Fc omit maps by blue meshes at the 3σ contour level. Substrates are shown as white sticks. (left) The biological assembly is identical with molecules in an asymmetric unit (RSMD: 0.170 Å). (right) A plausible bioassembly of EcOpgG in solution is a dimer, according to PISA analysis32. Chains A and B are a symmetry mate. c Superposition between the ligand-free and the Michaelis complex structure of EcOpgD (left) and EcOpgG (right) around the Loop A region. The ligand-free structures are shown in light yellow. (left) P78, N82, and W441 in the complex structure are shown as sticks. (right) Y56, S415, and L417 in the complex structure are shown as sticks.