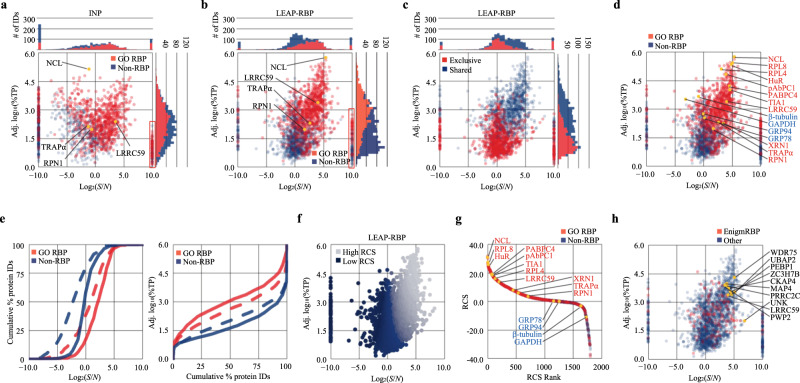
Fig. 6. High method specificity for RNA-bound RBPs allows accurate RCS ranking of RNA-binding proteins.
I INP, L LEAP-RBP, SILAC SILAC LC–MS/MS analysis, SRA SRA analysis. Specificity of I (a) and L (b) methods for enrichment of RNA-bound RBPs was evaluated by comparing observed abundances of RBPs and non-RBPs as a function of their log2(S/N) ratios (SILAC). Distributions of immunoblot targets reported in Fig. 5f demonstrates significance of vertical intercept. c L exclusives fall within the lower range of detection (%TP range). d, b With labeled immunoblot targets; red font: RNase-sensitive RBP; blue text: undetected or RNase-insensitive protein. e Cumulative frequency curves for RBPs (red) and non-RBPs (blue) identified in I (dashed) and L (solid) fractions as a function of log2(S/N) or log10(%TP) (SILAC). Effect of GO-annotation (GO:RBP) on analyzed using independent Kruskal–Wallis tests: significant effect of GO-annotation on log2(S/N) ratios of proteins identified in I and L fractions (SILAC), H(1) = 194.63, P < 0.001, H(1) = 436.62, P < 0.001, respectively; significant effect of GO-annotation on log10(%TP) of proteins identified in I and L fractions (SILAC), H(1) = 111.11, P < 0.001, H(1) = 632.29, P < 0.001, respectively. f, b With color overlay based on ordinal RCS rank. g RCS as a function of RCS rank. Protein IDs represent proteins examined by SRA and immunoblot in Fig. 2b; red font: RNase-sensitive RBP; blue font: undetected or RNase-insensitive proteins with positive RCS (i.e., log2(S/N) > 0) are considered more representative of their RNA-bound counterparts. h, f With top 10 RCS ranked enigmRNPs. Source data are provided as a Source Data file.