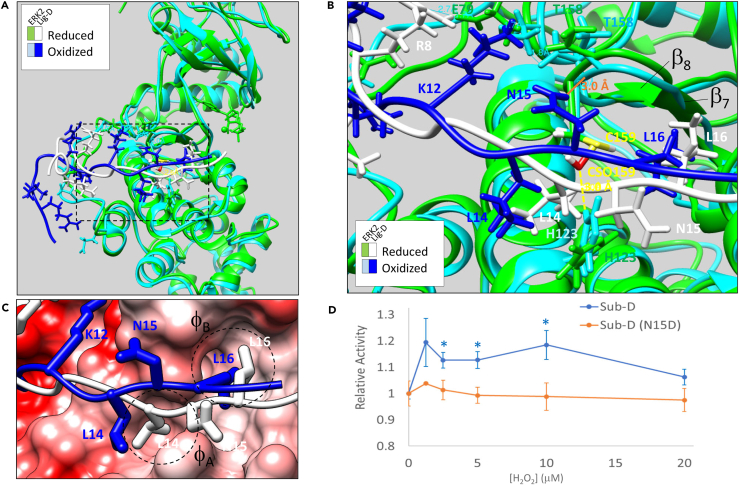
Figure 4.
Computational Modeling and Biochemical Investigation of ERK2-Lig-D Interactions
(A) Overlay of the final trajectories of 100 ns molecular dynamics simulations of the ERK2-Lig-D binding interaction with ERK2 in the oxidized (ERK2 in cyan; Lig-D in royal blue) and reduced (ERK2 in green; Lig-D in white) states.
(B) Close-up of the region boxed in A. Key residues in ERK2 and Lig-D are labeled in the color corresponding to the respective model. An H-bond that forms between Lig-DN15 and the backbone carbonyl of ERK2T158 in the oxidized state is shown (solid orange line). Likewise, the distance between the sulfenylated form of C159 (CSO159) and ERK2H123 in the oxidized state is shown (dashed yellow line).
(C) Coulombic surface representation of ERK2-Lig-D showing the positions of Lig-DL14 and Lig-DL16 relative to hydrophobic pockets A and B (φA and φB, respectively) in the DRS.
(D) Relative activity of ERK2 toward Sub-D (blue) or Sub-D(N15D) following oxidation of ERK2 with the indicated concentrations of H2O2. Data points and error bars are as in Figure 2E. Statistically significant differences between ERK2 activity toward Sub-D and Sub-D(N15D) are indicated by an asterisk (∗, p < 0.05). All structure images were generated using UCSF Chimera software (https://www.cgl.ucsf.edu/chimera/).