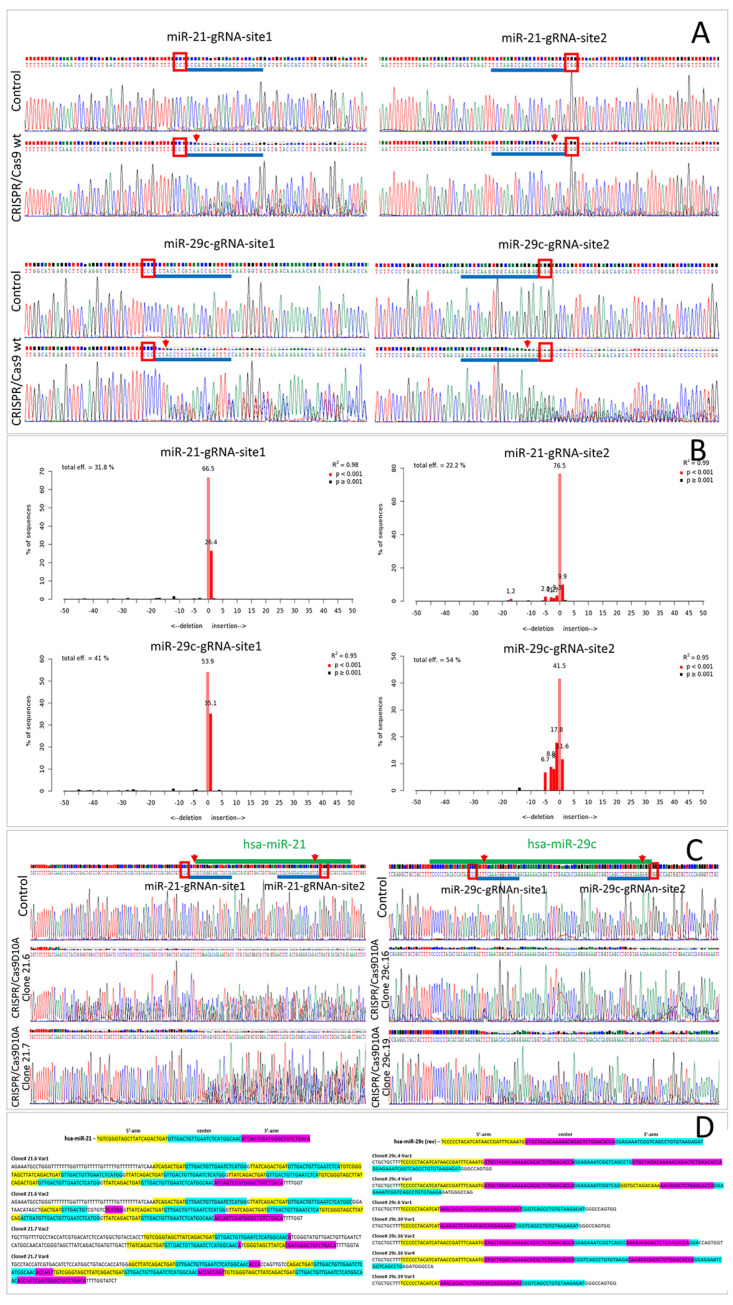
Figure 1.
Results of genomic DNA sequencing within the edited regions of ASC52telo cells. (A)—genome editing using CRISPR/Cas9 wild-type system (indels are visible, but no deletion of the intersite DNA sequence occurs); (B)—the results of analysis of edited loci using TIDE (CRISPR/Cas9 wild type); (C)—genome editing using CRISPR/Cas9 nickase system: clones 21.6, 21.7, 29c.16 and 29c.19 (indels are visible; however, it is impossible to establish the exact sequences of individual DNA loci); (D)—sample sequences of individual DNA loci within the hsa-mir-21 and hsa-mir-29c genes of clones 21.6, 21.7, 29c.4, 29c.10, 29c.16 and 29c.19, edited with the CRISPR/Cas9D10A nickase system. Red boxes (A,C)—PAM-sequences; a blue line next to red box (A,C)—gRNA protospacer; red arrows (A,C)—intended cut sites; light red column with the number above it (B)—the percentage of intact alleles; dark red column with the number above it (B)—the percentage of modified alleles (insertions or deletions).