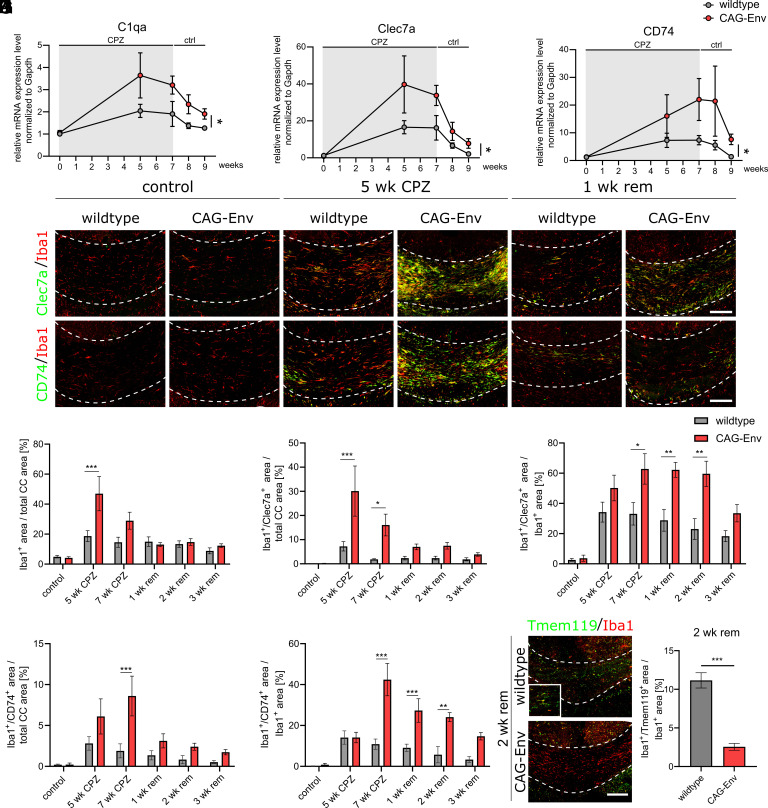
Fig. 3.
Transgenic HERV-W ENV expression activates microglial cells upon CPZ treatment. (A–C)C1qa, Clec7A, and CD74 gene expression analysis in the corpus callosum before and during CPZ treatment in wt vs. transgenic CAG-Env mice. (D) Representative immunohistochemical pictures of Clec7a/Iba1- and CD74/Iba1 coexpressing cells in wt and transgenic corpus callosum tissues (unchallenged and CPZ-treated animals). (E) Quantification of the Iba1-positive area over the total corpus callosum area in control vs. CPZ-treated wt and CAG-Env mice. (F) Quantification of Clec7a/Iba1 double-positive areas over total corpus callosum areas in wt and transgenic CAG-Env mice (control- and under CPZ diet). (G) Quantification of the proportion of Clec7a-positive microglia in wt and transgenic mice (control and under CPZ diet). (H) Quantification of CD74-positive microglial vs. total corpus callosum areas in wt and CAG-Env mice (control and under CPZ diet). (I) Quantification of the proportion of CD74-positive microglia in wt and transgenic mice (control- and under CPZ diet). (J) Representative immunohistochemical images of Tmem119/Iba1 coexpressing cells in wt and transgenic corpus callosum tissues at 2 wk of remyelination. (K) Quantification of the proportion of Tmem119-positive microglia in wt and transgenic mice at 2 wk of remyelination. Data are presented as mean values (n = 6) ± SEM. Significance of gene expression analysis was assessed by a Student’s unpaired t test of calculated AUCs whereas statistical significance of histological data was analyzed via 2-way ANOVA followed by Sidak’s post hoc test. Data were considered as statistically significant (95% CI) at *P < 0.05, **P < 0.01, ***P < 0.001. CC = corpus callosum. Dashed lines in (D and J) demarcate the corpus callosum. (Scale bar in D and J: 100 µm.)