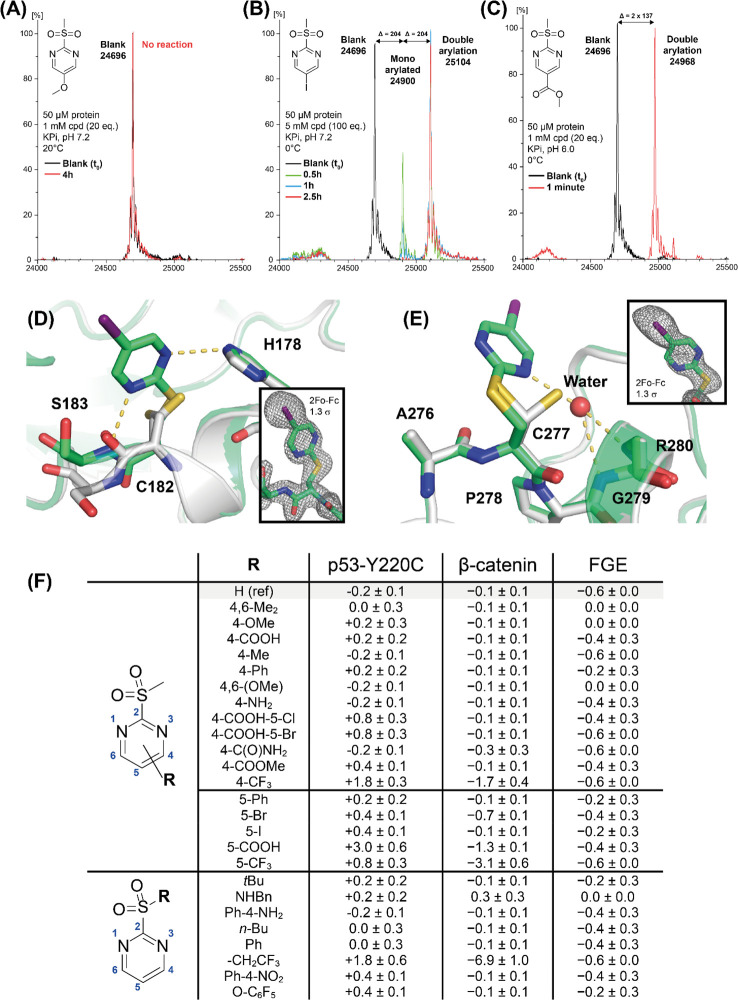
Figure 4.
Biophysical and structural characterization of protein cysteine arylation by 2-SPs. Top: Representative deconvoluted ESI (ES+) mass spectra of 50 μM p53-Y220C incubated with arylating agents 4n (A), 4u (B), 4y (C) (green/red) versus without compound (black) in KPi buffer. Stoichiometry, reaction time/temperature, and pH are indicated in each case. X axis: m/z (Da). Y axis: normalized intensities as a percentage of the most intense peak. Middle: Structure of the modified Y220C mutant (green) superimposed onto the structure of the unmodified protein (gray, PDB entry 6SHZ)40 showing the region around modified C182 (E) and C277 (F). Hydrogen bonds formed by the pyrimidine are highlighted as dashed yellow lines. 2Fo – Fc electron density maps are shown at a contour level of 1.3σ for segments of chain B including the modified residues C182 and C277. Bottom (F): Protein thermal stability (ΔTm, °C) of p53-Y220C, β-catenin ARD. and FGE, determined by DSF in the presence of 100 μM 2-SP derivatives.