Figure 8. R-state inhibitors promote outward movements of N-lobe structural elements.
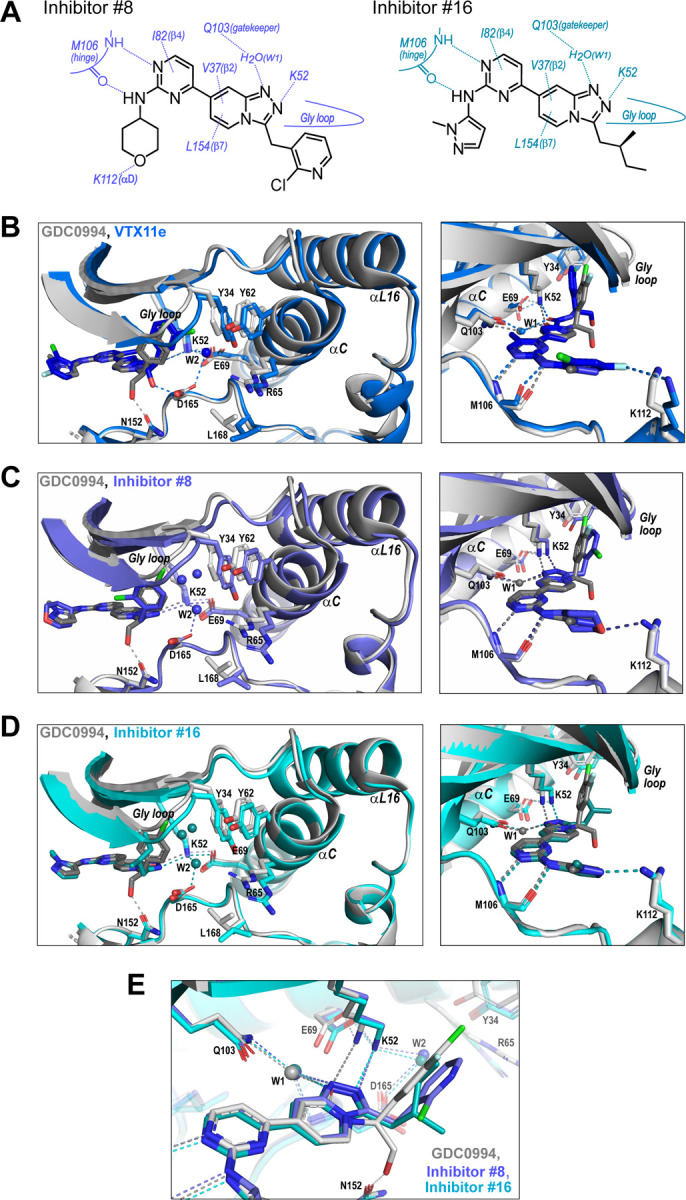
(A) Summary of contacts formed by inhibitors #8 and #16 with active site residues in co-crystal structures with 2P-ERK2. (B) The active site of 2P-ERK2 complexed with GDC0994 (PDBID:6OPH, grey) and VTX11e (PDBID:6OPK, blue) (Pegram et al, 2019). Left panel: Front view showing movement of the Gly loop, helix αC and helix αL16 in an “outward” direction (away from the inhibitor) upon binding of VTX11e (blue), relative to GDC0994 (grey). The movement can be attributed to the right-side 3-chlorobenzyl substituent in VTX11e which interacts with the π orbital of Y34 in the Gly loop (Cl-π distance, 3.5 Å). In turn, π–π stacking interactions between Y34 and Y62 couples movements of the Gly loop to helix αC. Right panel: Side view showing left-side hydrogen bond contacts with main chain atoms of hinge residue M106, as typical of ATP-competitive kinase inhibitors. (C,D) Active site of 2P-ERK2 complexed with GDC0994 (grey) and (C) inhibitor #8 (slate) or (D) inhibitor #16 (cyan). Like VTX11e, inhibitors #8 and #16 move the Gly loop, helix αC and helix αL16 outward, relative to GDC0994. Left panels B-D show that all inhibitor complexes share a bound water (W1) bridging the central scaffold to the gatekeeper residue in ERK2 (Q103). Right panels show that a bound water (W2) bridges active site residues K52 and D165 in complexes with VTX11e, inhibitor #8 and inhibitor #16, but not GDC0994. (E) Overlay of GDC0994 (grey), inhibitor #8 (slate) and inhibitor #16 (cyan). The relative N-lobe movements in panels C and D may be explained by differential hydrogen bonding of K52 and W1 to the triazolopyridine central scaffold of inhibitors #8 and #16, distinct from the pyridone scaffold of GDC0994. The position of the hydrogen bond of the triazole nitrogen with K52 relative to the pyridone oxygen moves the K52-E69 salt bridge in an outward direction in inhibitors #8 and #10 relative to GDC0994. Structures were superpositioned by aligning Cα atoms within the C-terminal domain (residues 109–141, 205–245, 272–310).
