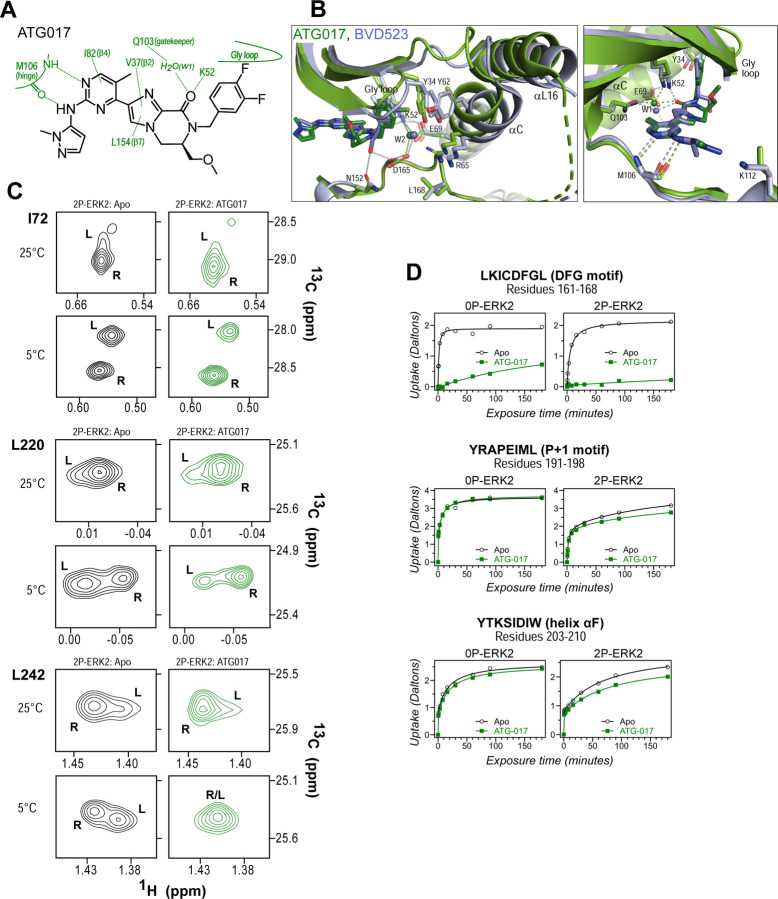
Figure 9. ATG017 promotes Gly loop opening and inward movement of N-lobe elements.
(A) Summary of contacts formed by inhibitor ATG017 with active site residues in a co-crystal structure with 0P-ERK2 (Ward et al., 2019). (B) The active site of 0P-ERK2 complexed with ATG017 (PDBID:6SLG, green) and BVD523 (PDBID:6GDQ, light blue) (Ward et al., 2019, Heightman et al, 2018). Left panel: Front view showing movement of the Gly loop, helix αC and helix αL16 in an outward direction by BVD523, attributed to the close proximity between the right-side chlorobenzyl substituent and the π orbital of Y34 in the Gly loop (Cl-π distance, 3.3 Å). Right panel: View showing left-side hydrogen bonds with main chain atoms of hinge residue M106. (C) 2D-HMQC spectra of 2P-ERK2 complexed with ATG017 at 25°C and 5°C, showing [methyl 1H,13C] peaks of residues I72, L220 and L242. Unlike 2P-ERK2 complexed with BVD523, the ATG017 complex retains R⇌L exchange resembling that of GDC0994 (Fig. 1). (D) HDX time courses with ATG017 measuring deuterium uptake at the DFG motif, P+1 segment, and helix αF. Time courses for strand β9 are shown in Suppl. Fig. S8. Enhanced HDX protection by ATG017 binding is observed at the DFG and adjacent β9 segments, but minimally at the P+1 and helix αF, similar to that seen with GDC0994. The results suggest that allosteric coupling between the ligand binding pocket and distal regions surrounding the activation loop, but not the DFG motif or β9, are characteristic of R-state inhibitors. Structures were superpositioned by aligning Cα atoms within the C-terminal domain (residues 109–141, 205–245, 272–310).