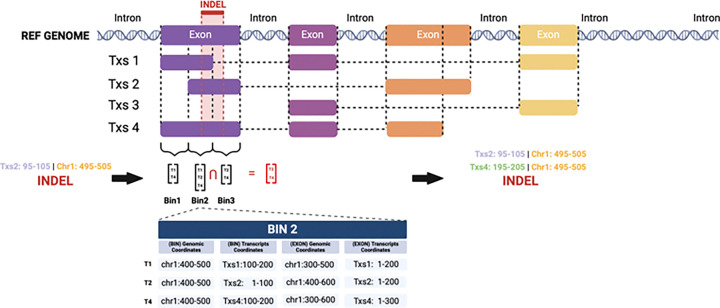
Figure 4. Schematic for Determining Equivalence Classes of Transcripts for INDEL variants.
To identify equivalence classes of transcripts for INDEL variants, the process beings by disjoining exon regions for each gene in the GFF3 file based on their genomic coordinates. For instance, a single exon may be split into multiple disjoint bins. In the provided example, the first exon generates three bins: Bin1 [T1, T2], Bin2 [T1, T2, T3], and Bin3 [T2, T3]. For each bin, the genomic and transcript-level coordinates of both the bin and the corresponding exon are tabulated (e.g., see the detailed example for Bin2). When analyzing an INDEL, all bins overlapping with the variant’s genomic coordinates are identified. Finally, the transcripts containing the INDEL are determined by intersecting the sets of transcripts associated with the overlapping bins.