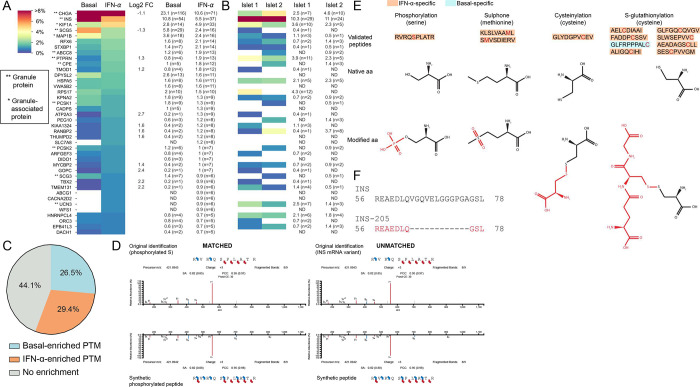
Figure 2. Immunopeptidome of ECN90 β-cells exposed or not to IFN-α and validation of HLA-I-eluted candidate neo-epitopes.
A-B. Heatmap of the relative representation of the top 40 source proteins in ECN90 β-cells (A) and human islets (B), ranked according to the number of peptides detected in the IFN-α-treated condition. The color scale is proportional to the number of peptides identified for each protein out of the total number of peptides in a given condition, expressed as percentage. Only conventional and PTM sequences are included. Peptides carrying PTMs were counted as such only for PTMs defined as likely biological (they were otherwise counted as unmodified); cis-spliced peptides were excluded. Percent values and peptide numbers are listed on the right. Source proteins enriched in IFN-α-treated cells (log2 fold change, FC ≥ 1) or in basal condition (log2 FC ≤ 1) are indicated for ECN90 β-cells. The complete heatmap is provided in Supplementary Fig. 2. HLA-I-bound peptides eluted from primary human islets are listed in Supplementary Data 5 and compared with those eluted from ECN90 β-cells in Supplementary Data 6. C. Global PTM enrichment in basal and IFN-α-treated condition. A detailed list is provided in Supplementary Data 7. D. Spectral matching examples of matched (left) and unmatched (right) synthetic peptides compared to the initial peptide identification using an online tool (https://www.proteomicsdb.org/use). E. Validated post-translationally modified peptides and representation of the native and modified aa. The PTM is indicated in red. A detailed list is provided in Supplementary Data 3. F. Peptide alignment of INS and INS-205 mRNA variant. The sequence of the HLA-eluted variant is indicated in red and correspond to a sequence spliced out as compared to the canonical INS mRNA.