Figure 1.
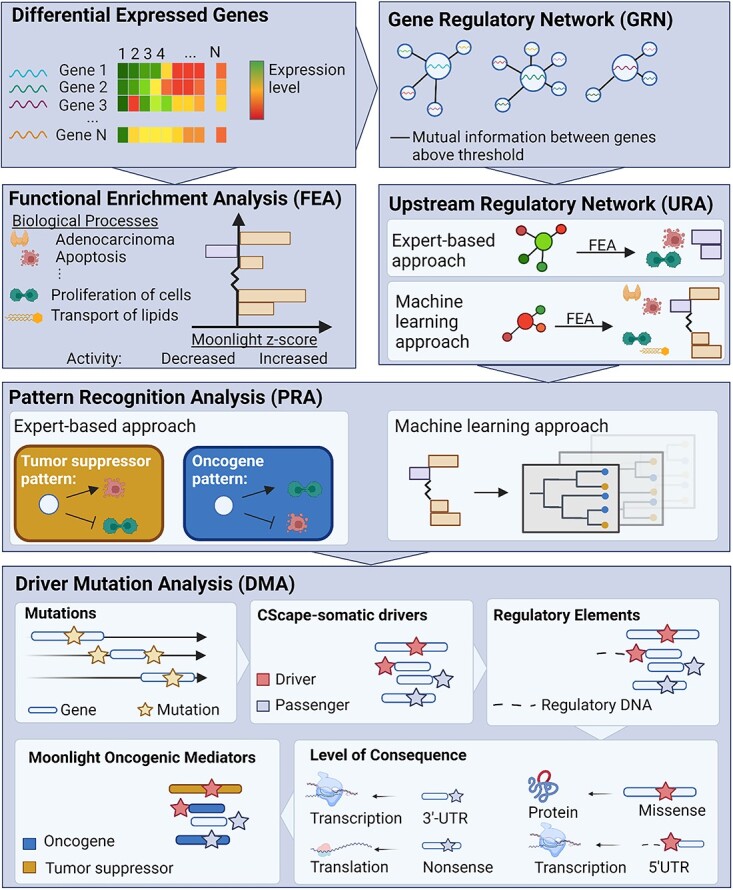
The Moonlight pipeline. The first step in Moonlight is a FEA which determines if any of Moonlight’s 101 cancer-related biological processes are enriched among an input set of differentially expressed genes (DEGs). This is done through Fisher’s exact tests and Moonlight Process Z-scores. The Moonlight Process Z-scores indicate if the activity of the process is increased or decreased based on literature reportings and gene expression levels. The next step, a gene regulatory network analysis (GRN), creates gene networks for each DEG by calculating the mutual information between all pairs of DEGs. Following GRN, the Moonlight pipeline diverges into an expert-based and a machine learning approach. The next step, an upstream regulatory analysis (URA), then evaluates the effect of each DEG on the biological processes through Moonlight Gene Z-scores. If the expert-based approach is selected, the Moonlight Gene Z-scores will only be calculated for chosen biological processes. If the machine learning approach is selected, the Moonlight Gene Z-scores will be calculated for all of Moonlight’s 101 biological processes. In the expert-based approach, pattern recognition analysis (PRA) then identifies the oncogenic mediators which fit an oncogene or tumor suppressor pattern based on the Moonlight Gene Z-scores. The two chosen biological processes must have opposite effects on cancer (growing/blocking), e.g. proliferation of cells and apoptosis. In the machine learning approach, the prediction of oncogenic mediators is done using a random forest classifier. Following Moonlight’s primary layer, a secondary mutational layer is applied through the DMA step which identifies driver mutations. First, the mutations are divided into passengers and drivers by CScape-somatic. Then, regulatory elements from ENCODE are added. The consequence of the type of mutation is annotated to either the protein’s structure, the level of transcription or level of translation. Finally, the data is cross-referenced with the Moonlight driver genes.
