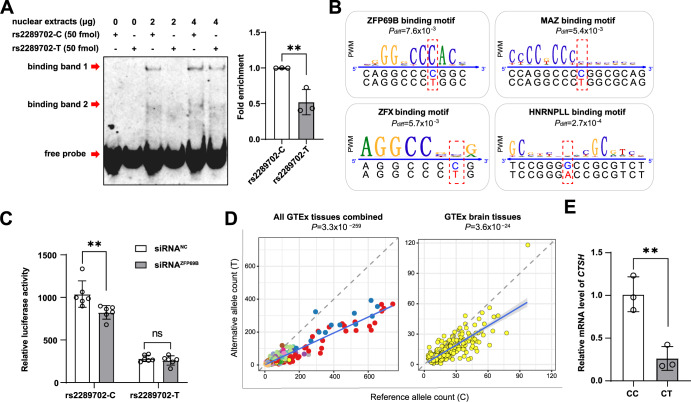
Fig. 2. SNP rs2289702 affects the binding affinity of transcription factors.
A SNP rs2289702 affected transcription factor binding affinity. The EMSA assays showed that allele C had a stronger binding affinity than allele T of rs2289702 in HEK293T cells (left) and quantification of the binding affinity (right). The arrows indicated two binding bands and the free probe. B Program affiliated prediction showing the location of rs2289702 in the binding motifs of transcription factors ZFP69B, ZFX, MAZ and HNRNPLL. The Pdiff value of the differential binding affinities of allele C and T of rs2289702 with each TF was computed by the atSNP algorithm [52]. C Knockdown of ZFP69B in human microglia (HM) cells affected the luciferase activity of the reporter vectors with different rs2289702 alleles. D Allele-specific expression of rs2289702 in GTEx human tissues [40]. The allele counts for the reference (Ref) allele and the alternative (Alt) allele were plotted, with each dot representing individual samples. Different tissues were marked by different colors. The blue line referred to the linear regression of all samples, and the gray shade represented the 95% confidence interval for the regression. The dashed line referred to the diagonal line. P value was measured by binomial tests. E Relative mRNA expression level of the CTSH gene in HEK293T cells with genotypes CC and CT of rs2289702. Values in A and E are presented as mean ± SD. ns not significant; *P < 0.05; **P < 0.01; two-tailed Student’s t test for A and E; two-way ANOVA test adjusted by Sidak’s multiple comparisons test for C.