Abstract
Telomere dysfunction is a notable event observed in many cancers contributing to their genomic instability. A major factor controlling telomere stability is the human telomerase reverse transcriptase catalytic subunit (hTERT). Telomere shortening has been observed in multiple myeloma (MM), a plasma cell malignancy with a complex and heterogeneous genetic background. In the present study, we aimed to analyse telomere length and hTERT genetic variants as potential markers of risk and survival in 251 MM patients. We found that telomere length was significantly shorter in MM patients than in healthy individuals, and patients with more advanced disease (stage III according to the International Staging System) had shorter telomeres than patients with less advanced disease. MM patients with hTERT allele rs2736100 T were characterized with significantly shorter progression-free survival (PFS). Moreover, allele rs2736100 T was also found to be less common in patients with disease progression in response to treatment. hTERT rs2853690 T was associated with higher haemoglobin blood levels and lower C-reactive protein. In conclusion, our results suggest that telomere length and hTERT genetic variability may affect MM development and can be potential prognostic markers in this disease.
Subject terms: Genetics, Cancer genetics, Clinical genetics, Epigenetics, Genetic markers, Genotype, Immunogenetics, Medical genetics, Cancer, Cancer genetics, Haematological cancer, Tumour biomarkers
Introduction
Multiple myeloma (MM) is an incurable haematologic malignancy, characterized by uncontrolled proliferation and accumulation of aberrant monoclonal plasma cells in the bone marrow1. The risk of developing MM increases with age, and it is more commonly diagnosed in males. The median age at diagnosis is 65 years, and the current mean survival of MM patients is approximately 5 years2. The symptomatic stage of MM is associated with lytic bone lesions, severe anaemia, hypercalcaemia, infections and kidney failure1. MM has a complex and heterogeneous genomic landscape. Its main feature is the presence of numerous genetic changes e.g. mutations, structural rearrangements and copy number variations3. Genomic alterations can be detected ranging from pre-malignant stages of monoclonal gammopathy of undetermined significance (MGUS) and smouldering MM (SMM) to clinical overt MM4.
Telomere dysfunction is one of the mechanisms that may lead to the genetic and clinical heterogeneity observed in MM. Telomeres are fragments of DNA located at the ends of linear chromosomes consisting of hexameric repeats of (5′-TTAGGG-3′)n. Chromosomes lacking telomeres may assemble abnormally and uncontrollably, causing genomic instability and changes in the karyotype5. In the group of MM patients, in addition to the shortening of telomere length with each cycle of cell proliferation, another important source of chromosomal instability was observed—telomere uncapping. This mechanism affects telomere structure and has been shown to be related to aggressiveness in MM at diagnosis6,7. Telomere length is tightly regulated by a reverse transcriptase called telomerase5. The primary task of telomerase is to extend the 3′ ends of chromosomes by adding short DNA fragments—telomeric repeats. Telomerase is a holoenzyme with reverse transcriptase properties, an integral component of which is the RNA template (TERC), and the human telomerase reverse transcriptase catalytic subunit (hTERT)8. Telomerase activity is suppressed in most human somatic cells and only retained in germ cells, activated T and B lymphocytes, and to some extent in stem cells9,10. Interestingly, telomerase activity is found in 90% of patients with newly diagnosed and relapsed MM. Short telomeres are observed in MM patients both at diagnosis and during disease progression. A study by Wu et al. showed that MM patients can be characterized by short telomere length and, at the same time, high levels of telomerase activity. The above observations support the concept of protection of critically short telomeric DNA by telomerase11. Nevertheless, it seems that the telomere shortening in haematological malignancies might still be associated with occurrence of aberrant karyotype12,13. In addition, inhibition of telomerase activity by GRN163L (Imetelstat), a lipid-conjugated thio-phosphoramidate oligonucleotide, was shown to be effective in the treatment of MM patients in both in vitro and in vivo studies14. Short telomeres may be responsible for chemotherapy resistance, as telomere shortening can result in adaptation of malignant cells, allowing their increased proliferation to overcome the cytotoxicity of treatment15.
The occurrence of telomere shortening also depends on the expression level, promoter mutations and genetic variability within the gene coding for hTERT16,17. The hTERT gene is located on the shorter arm of chromosome 5 (5p15.33) and consists of 15 introns, 16 exons and a 260 bp promoter core18. Aberrant gene variants may play a key role in transforming somatic cells into malignant cells by activating telomerase and other related signalling pathways, e.g. Wnt/β-catenin pathway19,20.
In the present study, we aimed to assess telomere length and hTERT genetic variants as potential markers associated with risk, survival, response to treatment and clinical course of the disease in MM. For this purpose, telomere length as well as six SNPs (rs2853690, rs2736100, rs33954691, rs35033501, rs2735940, rs10069690) located within the hTERT gene were analysed.
Results
Telomere length in multiple myeloma patients and healthy individuals
We observed that telomere length was significantly shorter in MM patients than in healthy individuals (median length 2.35 vs 4.96, p < 0.001, Fig. 1). This association was further confirmed in a multivariate generalized linear model analysis which included, alongside telomere length, also age and sex of patients and controls. Telomere length was proved to be an independent factor of MM risk (p = 0.010, Table 1).
Figure 1.
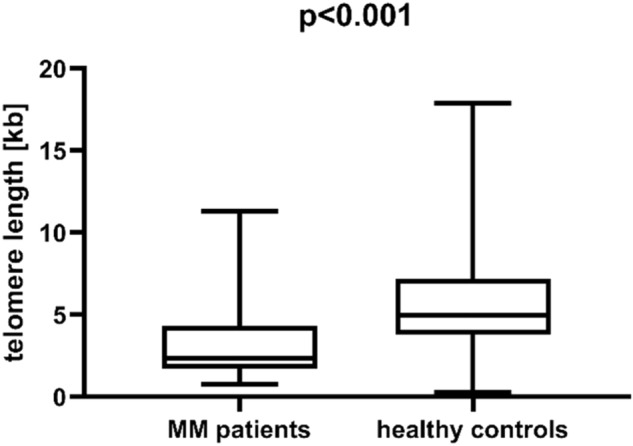
Telomere length in multiple myeloma (MM) patients and healthy individuals.
Table 1.
Results of a multivariate logistic regression analysis of multiple myeloma (MM) risk.
| Odds ratio (95% CI) | p-value | |
|---|---|---|
| Telomere length | 0.733 (0.579–0.928) | 0.010 |
| Age | 1.272 (0.193–1.356) | < 0.001 |
| Sex | 2.069 (0.683–6.264) | 0.198 |
Significant values are in bold.
Telomere length was also shorter in patients with more advanced disease (stage III according to the International Staging System; ISS) than in patients with less advanced disease (median length 2.27 vs 2.81, p = 0.031, Fig. 2). Nevertheless, patients with less advanced disease (ISS I-II) still had significantly shorter telomeres than healthy individuals (median length 2.81 vs 4.96, p < 0.001, Fig. 2). Telomere length also weakly correlated with blood albumin concentration (R = 0.293, p = 0.004). No associations with overall or progression-free survival or other clinical parameters was observed.
Figure 2.
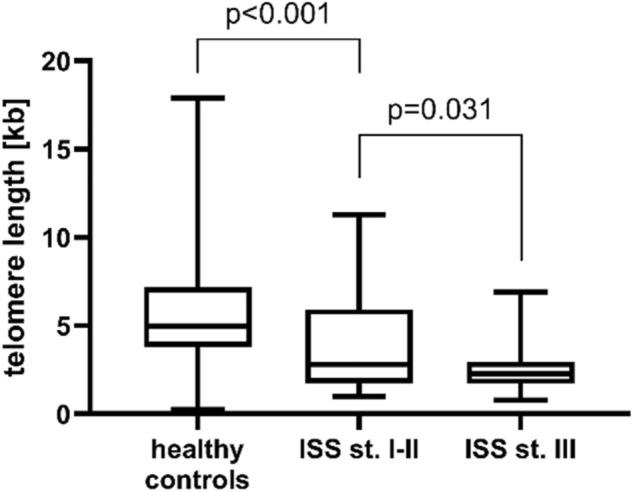
Telomere length in MM patients with less advanced (stages I–II according to the ISS) and more advanced (stage III) disease. Healthy individuals are also included for comparison. Patients with more advanced disease are characterized by shorter telomeres than those with less advanced disease, although both groups of patients have significantly shorter telomeres than healthy individuals.
hTERT genetic polymorphism and MM survival
Genotype frequencies for all the six analysed hTERT SNPs (rs2853690, rs2736100, rs33954691, rs35033501, rs2735940, rs10069690) are presented in Table 2. All the SNPs were in Hardy–Weinberg equilibrium in both patients and healthy controls. Linkage disequilibrium (LD) analysis showed that rs2736100, rs2735940, rs10069690 were in relatively high LD (Fig. 3). No difference was observed in the allele and genotype frequencies between MM patients and healthy individuals.
Table 2.
hTERT genotype frequencies in multiple myeloma (MM) patients and healthy individuals.
| MM patients (N = 251) | Healthy individuals (N = 226) | |
|---|---|---|
| rs2853690 | ||
| CC | 185 (73.7%) | 140 (71.8%) |
| CT | 62 (24.7%) | 55 (28.2%) |
| TT | 4 (1.6%) | 0 (0.0%) |
| rs2736100 | ||
| GG | 58 (23.1%) | 47 (20.8%) |
| GT | 118 (47.0%) | 115 (50.9%) |
| TT | 75 (29.9%) | 64 (28.3%) |
| rs33954691 | ||
| CC | 218 (86.9%) | 163 (84.0%) |
| CT | 32 (12.7%) | 28 (14.4%) |
| TT | 1 (0.4%) | 3 (1.6%) |
| rs35033501 | ||
| GG | 240 (95.6%) | 188 (96.4%) |
| GA | 11 (4.4%) | 7 (3.6%) |
| AA | 0 (0.0%) | 0 (0.0%) |
| rs2735940 | ||
| TT | 61 (24.3%) | 48 (21.3%) |
| TC | 125 (49.8%) | 111 (49.3%) |
| CC | 65 (25.9%) | 66 (29.3%) |
| rs10069690 | ||
| GG | 141 (56.2%) | 123 (54.4%) |
| GA | 93 (37.0%) | 89 (39.4%) |
| AA | 17 (6.8%) | 14 (6.2%) |
Figure 3.
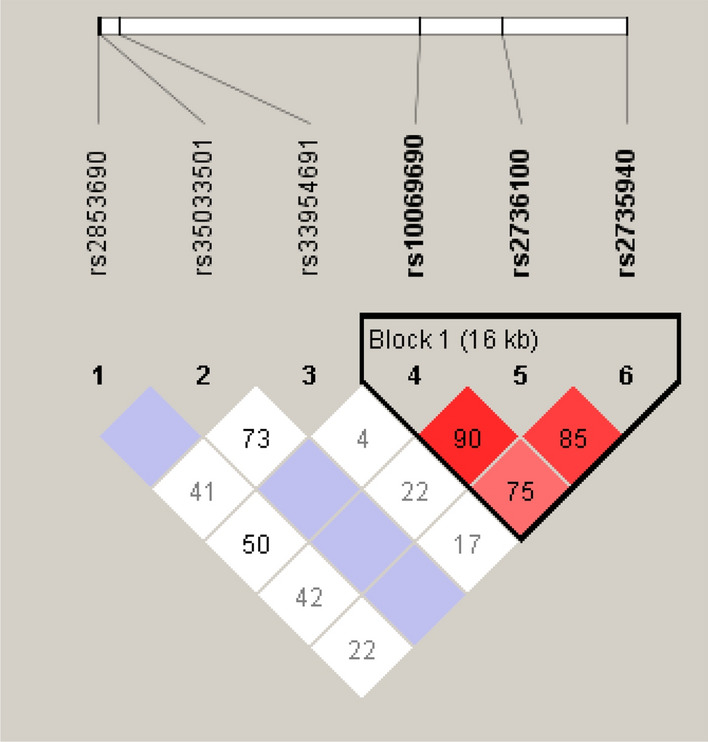
Linkage disequilibrium between hTERT SNPs included in the study. Darker colours represent higher D′ values. Results were obtained from the Haploview 4.2 software21.
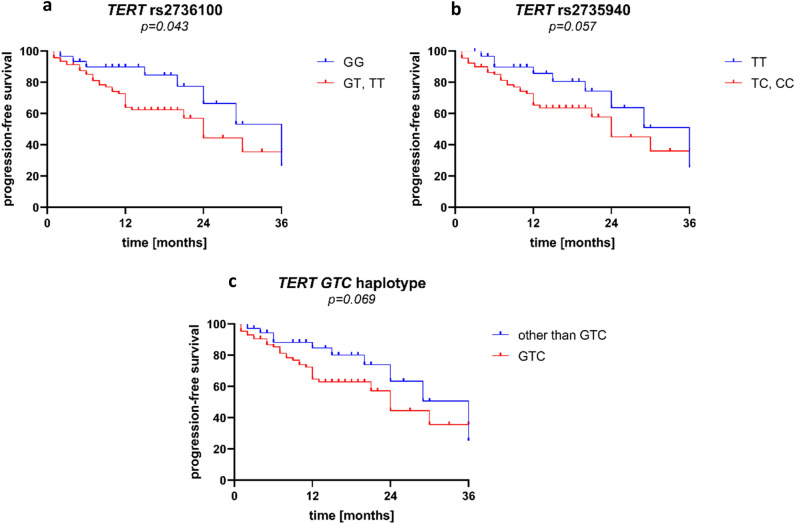
We performed an analysis of both overall (OS) and progression-free survival (PFS) to test if any of the hTERT SNPs might affect it. No association with OS was observed. We noticed that alleles rs2736100 T and rs2735940 C corresponded with shorter PFS during the first 3 years, but longer PFS thereafter (Supplementary Fig. S1). When analysing only early survival (first 3 years), patients with allele rs2736100 T were characterized with significantly shorter PFS than patients without this allele (p = 0.043, Fig. 4a). A similar association, although not statistically significant, was also observed for allele rs2735940 C (p = 0.057, Fig. 4b) and for the GTC (rs10069690 G, rs2736100 T, rs2735940 C, based on the LD data as shown in Fig. 3) haplotype (p = 0.069, Fig. 4c). These observations were confirmed in analyses in a Cox proportional hazards regression model including age and later ISS stages in addition to each of the risk alleles/haplotypes. Allele rs2736100 T (p = 0.035), but not allele rs2735940 C (p = 0.102) or haplotype GTC (p = 0.075), was found to be an independent factor of shorter PFS.
Figure 4.
Progression-free survival during the first 3 years in multiple myeloma patients and hTERT SNPs rs2736100 (a), rs2735940 (b), as well as the GTC (rs10069690 G, rs2736100 T, rs2735940 C) haplotype (c).
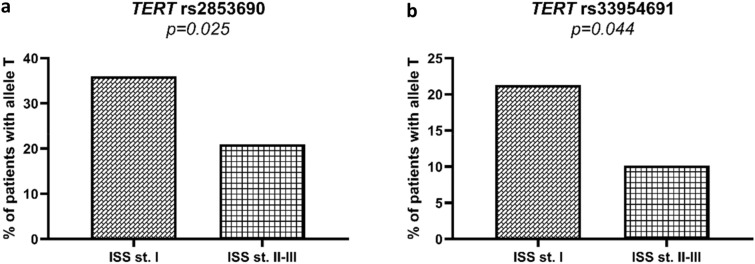
When stratified according to the ISS criteria, patients with less advanced disease (ISS I) on diagnosis more commonly carried the rs2853690 T allele than those diagnosed with more advanced (ISS II-III) disease (22/61 vs 35/167, p = 0.025, Fig. 5a). Similarly, patients with less advanced disease (ISS I) also carried allele rs33954691 T more commonly than other patients (13/61 vs 17/167, p = 0.044, Fig. 5b). Detailed information on distribution of hTERT genotypes in patients with different ISS stages is shown in Supplementary Table S1. Allele rs2853690 T was also associated with higher haemoglobin blood levels (median value 11.7 vs 10.7 g/dL; p = 0.006) and lower C-reactive protein (1.8 vs 4.6 mg/dL; p = 0.034) compared to patients without this allele. Interestingly, we also observed that alleles rs2736100 T, rs2735940 C, and haplotype GTC were less common in patients with progressive disease in response to treatment (p = 0.024, p = 0.020, p = 0.031 for the two alleles and the GTC haplotype, respectively).
Figure 5.
Frequency of alleles rs2853690 T (a) and rs33954691 T (b) among patients with more and less advanced disease at diagnosis according to the ISS criteria. Both alleles are more common in patients with ISS I.
We also analysed differences in telomere length between carriers of various hTERT alleles. MM patients with alleles rs10069690 G had longer telomeres than patients without this allele (2.33 vs 3.99 kb, p = 0.047). No associations with other SNPs included in this study were observed.
Discussion
The current knowledge of human cancer development shows that telomere dysfunction may be a key event causing genomic instability and disease progression in many types of solid tumours e.g. renal cell carcinoma22, glioma23,24, esophageal squamous cell carcinoma25, gastric cancer26, ovarian and breast cancer27–30. Additionally, telomere shortening has also been observed in haematological malignancies, both acute and chronic leukaemias31–34, some lymphomas35–37, as well as bone marrow failure syndromes (myelodysplastic syndrome (MDS))38,39 and aplastic anaemia40–42. It is not surprising that diseases related to telomere length manifest so prominently in the bone marrow, given that it is a place where turnover of cells is remarkably high, with app. 109 cells being produced every hour43. Haematopoiesis is based on the ability of stem cells to self-renewal and differentiation. In humans, there is clear evidence that short telomeres cause quantitative and qualitative defects in HSCs that manifest as stem cell depletion44–47.
MM is a very complex haematologic malignancy with a heterogenous genomic landscape, involving accumulation of mutations, as well as structural and copy number changes. These genetic variations occur in combination with point mutations and affect various cellular pathways, including genome maintenance3. Telomere dysfunction is one of the mechanisms that may lead to the genetic and clinical heterogeneity observed in MM, therefore analysis of telomere length may have prognostic significance. Our present study was conducted on a group of newly diagnosed untreated MM patients. We observed that significant shortening of telomere length in MM patients compared to healthy individuals. This is contrast to a study by Campa et al., as they found a link between longer telomeres and an increased risk of MM48. However, most other studies seem to confirm our observation. Similar results were reported by Wu et al. in a study analysing CD138 positive cells isolated from the bone marrow of patients with newly diagnosed or relapsed MM and healthy donors. These results showed significantly reduced telomere length in MM patients compared to telomere length in plasma cells from healthy donors11. Cottliar et al. studied bone marrow cells from patients with MM and with MGUS. Similarly to our results, they observed short telomeres in MM patients at diagnosis and during relapse. Additionally, they noted that telomere length in bone marrow cells was restored after disease remission49. In a three-dimensional (3D) telomere analysis, Klewes et al. showed changes in 3D nuclear architecture during disease progression from MGUS to MM, resulting in increased telomere attrition and consequent shorter telomeres in MM, as well as in relapsed MM, compared to MGUS50.
Hyatt et al. stratified their group of MM patients using a length threshold for telomere dysfunction (telomere length below 3.81 kb) previously defined by Lin et al.51,52. They showed that MM patients with short telomeres (< 3.81 kb) had significantly shorter overall survival. In their study, short telomeres were also associated with significantly worse survival in high-risk ISS patients. Moreover, in a multivariate modelling analysis, telomere length as well as patient’s age and ISS were identified as predictors of MM risk. Based on these data, Hyatt et al. proposed that each ISS prognostic subset could be further stratified for risk according to telomere length, supporting the inclusion of this parameter as a refinement of the ISS system51.
Telomere length can be influenced by genetic variability, including mutations in the telomerase components hTERT and TERC, and various single-nucleotide polymorphisms (SNPs), some of which are located near genes with known roles in telomere maintenance53. In addition, SNPs can affect telomere length, as their variants can modulate the expression of a broad spectrum of genes e.g. Myc, TP53 and NFKB30,54,55. For example, − 1327C>T (rs2735940) is a SNP located in the promoter region of hTERT gene (hTERTp) and is a T/C transition 1327-bp upstream of the transcription start site and is able to induce expression of hTERT by upregulating its transcriptional activity in vitro and hTERT mRNA expression in vivo53. This SNP has been extensively analysed in various types of solid cancers. However, it has not been studied much in haematological diseases, except for one study on patients with childhood ALL56. In this study, we observed that MM patients carrying C allele hTERTp rs2735940 had shorter PFS during the first 3 years. Sheng et al. reported that the TT genotype and T allele were associated with ALL in Chinese children56. However, conflicting results were obtained by Eskandari et al., who concluded that rs2735940 does not increase the risk of ALL57. In patients with renal cell carcinoma, the CC genotype hTERTp rs2735940 was associated with shorter time to disease progression and shorter overall survival58. In our present study, we did not observe any significant associations between rs2735940 and telomere length. However, there are reports suggesting that hTERT rs2735940 polymorphism affects telomere length and that longer telomeres were associated with an increased breast cancer (BC) and lung cancer risk59,60. Our previous study showed that women with BC and the CC genotype had longer telomeres than those with TC and TT genotypes. Additionally, BC patients with the rs2735940 C allele were characterized by more invasive tumours than patients with the TT genotype30.
The next SNP, rs2736100, is located in intron 2 of the hTERT gene and based on the result of the Evolutionary and Sequence Pattern Extraction through Reduced Representation score, it is located within a putative regulatory region61,62. It is the most frequently analysed polymorphic variant of hTERT. The C allele of rs2736100 was found to be associated with longer telomeres, which is consistent with the direct regulatory effect of the rs2736100 genotype on hTERT gene expression29. Interestingly, the effect strength of the rs2736100 polymorphism may vary between populations, as demonstrated, e.g. in Swedish and Chinese males with myeloproliferative neoplasms (MPNs)63. In addition, rs2736100 C is also associated with increased blood cell count, a hallmark of MPN64,65, and has been identified as a risk variant for MPN in the Icelandic population64. Tong et al. observed a higher frequency of the CC genotype and C allele in Chinese AML patients66. Furthermore, Rampazzo et al. observed longer telomeres at diagnosis and greater telomere erosion during neoadjuvant chemoradiotherapy in patients with rectal cancer with CC genotype compared to patients with the other genotypes67. In the present study, we observed that the T allele rs2736100 was associated with shorter early PFS in MM patients. We confirmed this observation (for T allele rs2736100) by a Cox proportional hazards regression model analysis, including age and later ISS stages in addition to each of the risk alleles. In our previous study, patients with CLL carrying C allele were characterized by longer telomeres with less advanced disease (Rai 0–I or Binet A) compared to patients with the C allele, but exhibiting more advanced stage of CLL32. In women with BC we showed that patients with T allele rs2736100 had more invasive tumours than BC patients with other genotypes30.
Another SNP, rs10069690, was much more extensively studied. It is known to act as a risk factor in many cancers68 and was found to be a marker of decreased risk for MM in an earlier study48. However, we were not able to confirm this association in our present study. Three other SNPs included in our study were predicted to affect either miRNA binding (rs2853690) or splicing (rs33954691, rs35033501) based on an analysis by the National Institute of Environmental Health Sciences SNP Function Prediction tool69. Of these, we found rs2853690 T and rs33954691 T to be more common in patients with less advanced disease (lower ISS stage). Additionally, rs2853690 T was also associated with higher haemoglobin and lower C-reactive protein levels, low haemoglobin and high CRP being common features of MM progression70,71. All three of those SNPs have been relatively poorly studied. The rs2853690 is known to affect circulating hTERT mRNA levels and response to treatment in patients with rectal cancer, as well as spontaneous preterm labour in pregnant women67,72. rs33954691 was shown to affect longevity and leucocyte telomere length73,74. It was also associated with increased risk of radioiodine-refractory papillary thyroid carcinoma75. All three SNPs were analysed in our previous study on chronic lymphocytic leukaemia (CLL), although only rs35033501 was associated with CLL risk32.
The regulation of the hTERT gene is a very complex process and hTERTp can be activated by multiple mechanisms in haematological malignancies. Moreover, genetic variation in hTERT may modulate telomere length and thus such genetic variants may be risk factors for cancer development. So, it appears that the optimal telomere length is a balance of cell proliferation, senescence and control. Shortening of telomeres to critical length results in a loss of telomere protection, leading to chromosomal instability, which can contribute to the abnormalities in the hematopoietic process.
Methods
Patients and controls
The study included 251 newly diagnosed Polish MM patients and 226 healthy blood donors serving as the control group. Both groups were nearly equally divided into men and women (the ratio of females was 0.502 and 0.420, respectively). Blood samples were collected at diagnosis after obtaining informed consent from patients. All methods were used according to the Declaration of Helsinki. The study was approved by the Wroclaw Medical University Bioethical Committee (ethical approval code: 369/2019). According to International Staging System (ISS) stratification, 61 (24.3%) patients were in stage I at diagnosis, 76 (30.3%) were in stage II, 91 (36.3%) were in stage III, and 23 lacked data on ISS. Most patients were administered either cyclophosphamide, thalidomide, dexamethasone (CTD)—38.8%, bortezomib, thalidomide, dexamethasone (VTD)—13.2%, or bortezomib, melphalan, prednisone (VMP)—11.0% as first line therapy. Response to treatment was as follows: complete response (n = 43), very good partial response (n = 32), partial response (n = 81), minor response (n = 7), stable disease (n = 20), progressive disease (n = 19).
DNA extraction
Genomic DNA was isolated from peripheral blood taken on EDTA using Maxwell 16 Blood DNA Purification Kit (Promega Corporation, Madison, WI, USA) and the Qiagen DNA Isolation Kit (Qiagen, Hilden, Germany) following the recommendation of the manufacturers. DNA concentration and purity were quantified on a DeNovix DS-11 spectrophotometer (DeNovix Inc., Wilmington, DE, USA). The isolated DNA was then stored at − 20 °C until hTERT genotyping and evaluation of the telomere length in MM patients and healthy individuals.
Genotyping of hTERT gene polymorphisms
The selection of the studied single nucleotide polymorphisms (SNPs) within the hTERT gene was based on results of the SNP Function Prediction tool available on the website of the National Institute of Environmental Health Sciences (NCBI Database), as well as other auxiliary databases (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html (accessed on 9 January, 2023); https://www.ncbi.nlm.nih.gov/snp/ (accessed on 9 January, 2023); https://www.ensembl.org/index.html (accessed on 9 January, 2023). The following criteria were used: minor allele frequency in Caucasians above 10%, change in RNA and/or amino acid chain, potential splicing site and/or miRNA binding site. One SNP (rs35033501) was additionally included based on results from previous studies32.
Based on the above criteria, six hTERT SNPs were selected for the study: rs35033501 (G>A) located in exon 16; rs33954691 (C>T) located in exon 14; rs2853690 (C>T) located in the 3ʹ untranslated region (3′UTR); rs10069690 (G>A) located in intron 4; rs2736100 (G>T) located in intron 2; rs2735940 (T>C) located in the promoter region − 1327 bp upstream of the transcription start site. The hTERT polymorphisms were determined by LightSNiP typing assays (TIB MOLBIOL, Berlin, Germany) using quantitative polymerase chain reaction (qPCR). Amplifications were performed on a LightCycler480 II Real-Time PCR system (Roche Diagnostics International AG, Rotkreuz, Switzerland) according to the recommendations of the manufacturer. The PCR conditions were as follows: 95 °C for 10 min followed by 45 cycles of 95 °C for 10 s, 60 °C for 10 s and 72 °C for 15 s. PCR was followed by one cycle of 95 °C for 30 s, 40 °C for 2 min and gradual melting from 75 to 40 °C.
Quantification of telomere length
Mean telomere length was measured in the genomic DNA samples of 112 MM patients and 185 healthy controls. The group was almost equally divided into men and women (the ratio of females was 0.455). According to ISS stratification, 29 (25.9%) patients were in stage I, 39 (34.8%) were in stage II, 43 (38.4%) were in stage III, and 1 lacked data on ISS. The DNA samples were diluted with nuclease-free water to a concentration of 5 ng/mL. Telomere length measurements were performed on a LightCycler480 II Real-Time PCR system (Roche Diagnostics International, Rotkreuz, Switzerland) using qPCR test kits (ScienCell’s Absolute Human Telomere Length Quantification qPCR Assay Kit [AHTLQ], Carlsbad, CA, USA), as previously described by Dratwa et al.76. The PCR conditions were as follows: 95 °C for 10 min followed by 32 cycles of 95 °C for 20 s, 52 °C for 20 s and 72 °C for 45 s. Data analysis was conducted according to the manufacturer’s instructions. All reactions were run in three replicates.
Statistical analysis
The null hypothesis that there is no difference between the frequency of alleles and genotypes between patients and controls was verified with the Fisher’s exact test, calculated using the online tool http://vassarstats.net/tab2x2.htm (version as of 3 April, 2023). Mann–Whitney U test and logistic regression model were used to compare telomere length between patients and controls, and to check for associations between various clinical parameters and presence of various genetic variants. Correlations between telomere length and clinical parameters were assessed by Spearman’s coefficient. Survival of patients was analysed using the Gehan-Breslow-Wilcoxon test and Kaplan–Meier curves, as well as the Cox proportional hazards model. All of these analyses were conducting using the Real Statistics Resource Pack for Microsoft Excel 2013 version 15.0.5023.1000 (Microsoft, Redmond, WA, USA), RStudio (RStudio, PBC., Boston, MA, USA), and GraphPad Prism (version 8.0.1, GraphPad Software, San Diego, CA, USA). P-values < 0.05 were considered statistically significant, while those between 0.05 and 0.10 were indicative of a trend.
Supplementary Information
Author contributions
M.D.: conceptualization, methodology, investigation, writing—original draft, writing—review and editing. P.Ł.: conceptualization, methodology, formal analysis, writing—original draft, writing—review and editing. A.B.: resources, data curation (patient recruitment, sample collection, provision of clinical data). D.P.: data curation (sample collection, provision of clinical data). G.M.: resources, data curation (patient recruitment, sample collection, provision of clinical data). K.B-K.: conceptualization, methodology, writing—review and editing, supervision, project administration and funding. All authors have read and agreed to the published version of the manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Marta Dratwa and Piotr Łacina.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-43141-7.
References
- 1.Gerecke C, Fuhrmann S, Strifler S, Schmidt-Hieber M, Einsele H, Knop S. The diagnosis and treatment of multiple myeloma. Dtsch. Arztebl. Int. 2016;113:470–476. doi: 10.3238/arztebl.2016.0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naymagon L, Abdul-Hay M. Novel agents in the treatment of multiple myeloma: A review about the future. J. Hematol. Oncol. 2016;9:52. doi: 10.1186/s13045-016-0282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aksenova AY, et al. Genome instability in multiple myeloma: Facts and factors. Cancers. 2021;13:5949. doi: 10.3390/cancers13235949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dutta AK, et al. Subclonal evolution in disease progression from MGUS/SMM to multiple myeloma is characterised by clonal stability. Leukemia. 2019;33:457–468. doi: 10.1038/s41375-018-0206-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uziel O, et al. BRCA1/2 mutations perturb telomere biology: Characterization of structural and functional abnormalities in vitro and in vivo. Oncotarget. 2016;7:2433–2454. doi: 10.18632/oncotarget.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rangel-Pozzo A, et al. Telomere architecture correlates with aggressiveness in multiple myeloma. Cancers. 2021;13:1969. doi: 10.3390/cancers13081969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandes SG, et al. Role of telomeres and telomeric proteins in human malignancies and their therapeutic potential. Cancers. 2020;12:1901. doi: 10.3390/cancers12071901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greider CW, Blackburn EH. The telomere terminal transferase of tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 9.Hiyama K, et al. Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J. Immunol. 1995;155:3711–3715. [PubMed] [Google Scholar]
- 10.Broccoli D, Young JW, de Lange T. Telomerase activity in normal and malignant hematopoietic cells. Proc. Natl. Acad. Sci. USA. 1995;92:9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu KD, Orme LM, Shaughnessy J, Jr, Jacobson J, Barlogie B, Moore MA. Telomerase and telomere length in multiple myeloma: Correlations with disease heterogeneity, cytogenetic status, and overall survival. Blood. 2003;101:4982–4989. doi: 10.1182/blood-2002-11-3451. [DOI] [PubMed] [Google Scholar]
- 12.Thomay K, et al. Telomere shortening, TP53 mutations and deletions in chronic lymphocytic leukemia result in increased chromosomal instability and breakpoint clustering in heterochromatic regions. Ann. Hematol. 2017;96:1493–1500. doi: 10.1007/s00277-017-3055-1. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann U, Brümmendorf TH, Balabanov S, Thiede C, Illme T, Schaich M. Telomere length and hTERT expression in patients with acute myeloid leukemia correlates with chromosomal abnormalities. Haematologica. 2005;90:307–316. [PubMed] [Google Scholar]
- 14.Shammas MA, et al. Telomerase inhibitor GRN163L inhibits myeloma cell growth in vitro and in vivo. Leukemia. 2008;22:1410–1418. doi: 10.1038/leu.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nogueira BMD, Machado CB, Montenegro RC, de Moraes MEA, Moreira-Nunes CA. Telomere length and hematological disorders: A review. In Vivo. 2020;34:3093–3101. doi: 10.21873/invivo.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lansdorp PM. Maintenance of telomere length in AML. Blood Adv. 2017;1:2467–2472. doi: 10.1182/bloodadvances.2017012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dratwa M, Wysoczańska B, Łacina P, Kubik T, Bogunia-Kubik K. TERT-regulation and roles in cancer formation. Front. Immunol. 2020;11:589929. doi: 10.3389/fimmu.2020.589929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akincilar SC, Unal B, Tergaonkar V. Reactivation of telomerase in cancer. Cell Mol. Life Sci. 2016;73:1659–1670. doi: 10.1007/s00018-016-2146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong T, Luo M, Liu Q. The TERT rs2736100 polymorphism and susceptibility to myeloproliferative neoplasms: A systematic review and meta-analysis. Genet. Test Mol. Biomarkers. 2020;24:181–187. doi: 10.1089/gtmb.2019.0277. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez RJ, III, Johnson FB. A regulatory loop connecting WNT signaling and telomere capping: Possible therapeutic implications for dyskeratosis congenita. Ann. N. Y. Acad. Sci. 2018;1418:56–68. doi: 10.1111/nyas.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Park J, et al. Telomere length in peripheral blood leukocytes and risk of renal cell carcinoma. Transl. Cancer Res. 2019;8(Suppl 4):S397–S403. doi: 10.21037/tcr.2019.06.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, et al. Association between leukocyte telomere length and glioma risk: A case-control study. Neuro Oncol. 2014;16:505–512. doi: 10.1093/neuonc/not240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, et al. Leukocyte telomere length: A novel biomarker to predict the prognosis of glioma patients. J. Cancer Res. Clin. Oncol. 2015;141:1739–1747. doi: 10.1007/s00432-015-1938-x. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, et al. Identification of leukocyte telomere length-related genetic variants contributing to predisposition of esophageal squamous cell carcinoma. J. Cancer. 2020;11:5025–5031. doi: 10.7150/jca.45165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu F, et al. Short telomere length in peripheral blood leukocyte predicts poor prognosis and indicates an immunosuppressive phenotype in gastric cancer patients. Mol. Oncol. 2015;9:727–739. doi: 10.1016/j.molonc.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirabello L, et al. Leukocyte telomere length in a population-based case-control study of ovarian cancer: A pilot study. Cancer Causes Control. 2010;21:77–82. doi: 10.1007/s10552-009-9436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terry KL, et al. Telomere length and genetic variation in telomere maintenance genes in relation to ovarian cancer risk. Cancer Epidemiol. Biomarkers Prev. 2012;21:504–512. doi: 10.1158/1055-9965.EPI-11-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bojesen SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 2013;45:371–384.e3842. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dratwa M, et al. Relationship between telomere length, TERT genetic variability and TERT, TP53, SP1, MYC gene co-expression in the clinicopathological profile of breast cancer. Int. J. Mol. Sci. 2022;23:5164. doi: 10.3390/ijms23095164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dratwa M, Wysoczańska B, Butrym A, Łacina P, Mazur G, Bogunia-Kubik K. TERT genetic variability and telomere length as factors affecting survival and risk in acute myeloid leukaemia. Sci. Rep. 2021;11:23301. doi: 10.1038/s41598-021-02767-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wysoczanska B, et al. Variability within the human TERT gene, telomere length and predisposition to chronic lymphocytic leukemia. Onco Targets Ther. 2019;12:4309–4320. doi: 10.2147/OTT.S198313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruedigam C, Lane SW. Telomerase in hematologic malignancies. Curr. Opin. Hematol. 2016;23:346–353. doi: 10.1097/MOH.0000000000000252. [DOI] [PubMed] [Google Scholar]
- 34.Keller G, Brassat U, Braig M, Heim D, Wege H, Brümmendorf TH. Telomeres and telomerase in chronic myeloid leukaemia: Impact for pathogenesis, disease progression and targeted therapy. Hematol. Oncol. 2009;27:123–129. doi: 10.1002/hon.901. [DOI] [PubMed] [Google Scholar]
- 35.Chevret E, et al. Telomerase functions beyond telomere maintenance in primary cutaneous T-cell lymphoma. Blood. 2014;123:1850–1859. doi: 10.1182/blood-2013-05-500686. [DOI] [PubMed] [Google Scholar]
- 36.Ropio J, Merlio JP, Soares P, Chevret E. Telomerase activation in hematological malignancies. Genes. 2016;7:61. doi: 10.3390/genes7090061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Machiela MJ, et al. Genetically predicted longer telomere length is associated with increased risk of B-cell lymphoma subtypes. Hum. Mol. Genet. 2016;25:1663–1676. doi: 10.1093/hmg/ddw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sashida G, et al. Telomere dynamics in myelodysplastic syndrome determined by telomere measurement of marrow metaphases. Clin. Cancer Res. 2003;9:1489–1496. [PubMed] [Google Scholar]
- 39.Colla S, et al. Telomere dysfunction drives aberrant hematopoietic differentiation and myelodysplastic syndrome. Cancer Cell. 2015;27:644–657. doi: 10.1016/j.ccell.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi H, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N. Engl. J. Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 41.Scheinberg P, Cooper JN, Sloand EM, Wu CO, Calado RT, Young NS. Association of telomere length of peripheral blood leukocytes with hematopoietic relapse, malignant transformation, and survival in severe aplastic anemia. JAMA. 2010;304:1358–1364. doi: 10.1001/jama.2010.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakaguchi H, et al. Peripheral blood lymphocyte telomere length as a predictor of response to immunosuppressive therapy in childhood aplastic anemia. Haematologica. 2014;99:1312–1316. doi: 10.3324/haematol.2013.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metcalf D. The Molecular Control of Blood Cells. Harvard Univ. Press; 1998. [Google Scholar]
- 44.Parry EM, Alder JK, Qi X, Chen JJ, Armanios M. Syndrome complex of bone marrow failure and pulmonary fibrosis predicts germline defects in telomerase. Blood. 2016;127:1837. doi: 10.1182/blood-2010-11-322149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allsopp RC, Morin GB, DePinho R, Harley CB, Weissman IL. Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood. 2003;102:517–520. doi: 10.1182/blood-2002-07-2334. [DOI] [PubMed] [Google Scholar]
- 46.Fogarty PF, et al. Late presentation of dyskeratosis congenita as apparently acquired aplastic anaemia due to mutations in telomerase RNA. Lancet. 2003;362:1628–1630. doi: 10.1016/S0140-6736(03)14797-6. [DOI] [PubMed] [Google Scholar]
- 47.Armanios M, Blackburn EH. The telomere syndromes. Nat. Rev. Genet. 2013;14:235. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campa D, et al. Risk of multiple myeloma is associated with polymorphisms within telomerase genes and telomere length. Int. J. Cancer. 2015;136:E351–E358. doi: 10.1002/ijc.29101. [DOI] [PubMed] [Google Scholar]
- 49.Cottliar A, Pedrazzini E, Corrado C, Engelberger MI, Narbaitz M, Slavutsky I. Telomere shortening in patients with plasma cell disorders. Eur. J. Haematol. 2003;71:334–340. doi: 10.1034/j.1600-0609.2003.00157.x. [DOI] [PubMed] [Google Scholar]
- 50.Klewes L, et al. Three-dimensional nuclear telomere organization in multiple myeloma. Transl. Oncol. 2013;6:749–756. doi: 10.1593/tlo.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyatt S, et al. Telomere length is a critical determinant for survival in multiple myeloma. Br. J. Haematol. 2017;178:94–98. doi: 10.1111/bjh.14643. [DOI] [PubMed] [Google Scholar]
- 52.Lin TT. Telomere dysfunction accurately predicts clinical outcome in chronic lymphocytic leukaemia, even in patients with early stage disease. Br. J. Haematol. 2014;167:214–223. doi: 10.1111/bjh.13023. [DOI] [PubMed] [Google Scholar]
- 53.Matsubara Y, et al. Telomere length of normal leukocytes is affected by a functional polymorphism of hTERT. Biochem. Biophys. Res. Commun. 2006;341:128–131. doi: 10.1016/j.bbrc.2005.12.163. [DOI] [PubMed] [Google Scholar]
- 54.Codd V, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 2013;45:422–427.e4272. doi: 10.1038/ng.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delgado DA, et al. Genome-wide association study of telomere length among South Asians identifies a second RTEL1 association signal. J. Med. Genet. 2018;55:64–71. doi: 10.1136/jmedgenet-2017-104922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheng X. TERT polymorphisms modify the risk of acute lymphoblastic leukemia in Chinese children. Carcinogenesis. 2013;34:228–235. doi: 10.1093/carcin/bgs325. [DOI] [PubMed] [Google Scholar]
- 57.Eskandari E, Hashemi M, Naderi M, Bahari G, Safdari V, Taheri M. Leukocyte telomere length shortening, hTERT genetic polymorphisms and risk of childhood acute lymphoblastic leukemia. Asian Pac. J. Cancer Prev. 2018;19:1515–1521. doi: 10.22034/APJCP.2018.19.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morais M, et al. Leukocyte telomere length and hTERT genetic polymorphism rs2735940 influence the renal cell carcinoma clinical outcome. Future Oncol. 2020;16:1245–1255. doi: 10.2217/fon-2019-0795. [DOI] [PubMed] [Google Scholar]
- 59.Pellatt AJ, et al. Telomere length, telomere-related genes, and breast cancer risk: The breast cancer health disparities study. Genes Chromosomes Cancer. 2013;52:595–609. doi: 10.1002/gcc.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seow WJ, et al. Telomere length in white blood cell DNA and lung cancer: A pooled analysis of three prospective cohorts. Cancer Res. 2014;74:4090–4098. doi: 10.1158/0008-5472.CAN-14-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor J, Tyekucheva S, King DC, Hardison RC, Miller W, Chiaromonte F. ESPERR: Learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mocellin S, et al. Telomerase reverse transcriptase locus polymorphisms and cancer risk: A field synopsis and meta-analysis. J. Natl. Cancer Inst. 2012;104:840–854. doi: 10.1093/jnci/djs222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dahlström J, et al. TERT rs2736100 genotypes are associated with differential risk of myeloproliferative neoplasms in Swedish and Chinese male patient populations. Ann. Hematol. 2016;95:1825–1832. doi: 10.1007/s00277-016-2787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oddsson A, et al. The germline sequence variant rs2736100_C in TERT associates with myeloproliferative neoplasms. Leukemia. 2014;28:1371–1374. doi: 10.1038/leu.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamatani Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 66.Tong Y, et al. Association between TERT gene polymorphisms and acute myeloid leukemia susceptibility in a Chinese population: A case-control study. Cancer Cell Int. 2020;20:313. doi: 10.1186/s12935-020-01335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rampazzo E, et al. Genetic variants of the TERT gene, telomere length, and circulating TERT as prognostic markers in rectal cancer patients. Cancers. 2020;12:3115. doi: 10.3390/cancers12113115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He G, et al. TERT rs10069690 polymorphism and cancers risk: A meta-analysis. Mol. Genet. Genomic Med. 2019;7:e00903. doi: 10.1002/mgg3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Z, Taylor JA. SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Birgegård G, Gascón P, Ludwig H. Evaluation of anaemia in patients with multiple myeloma and lymphoma: Findings of the European Cancer Anaemia Survey. Eur. J. Haematol. 2006;77:378–386. doi: 10.1111/j.1600-0609.2006.00739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang J, et al. C-reactive protein promotes bone destruction in human myeloma through the CD32-p38 MAPK-Twist axis. Sci. Signal. 2017;10:eaan6282. doi: 10.1126/scisignal.aan6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marrs C, Chesmore K, Menon R, Williams S. Maternal human telomerase reverse transcriptase variants are associated with preterm labor and preterm premature rupture of membranes. PLoS ONE. 2018;13:e0195963. doi: 10.1371/journal.pone.0195963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soerensen M, et al. Genetic variation in TERT and TERC and human leukocyte telomere length and longevity: A cross-sectional and longitudinal analysis. Aging Cell. 2012;11:223–227. doi: 10.1111/j.1474-9726.2011.00775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bai Y, et al. Co-exposure to multiple metals, TERT-CLPTM1L variants, and their joint influence on leukocyte telomere length. Environ. Int. 2020;140:105762. doi: 10.1016/j.envint.2020.105762. [DOI] [PubMed] [Google Scholar]
- 75.Shen CT, Zhang GQ, Qiu ZL, Song HJ, Sun ZK, Luo QY. Targeted next-generation sequencing in papillary thyroid carcinoma patients looking for germline variants predisposing to the disease. Endocrine. 2019;64:622–631. doi: 10.1007/s12020-019-01878-0. [DOI] [PubMed] [Google Scholar]
- 76.Dratwa M, et al. Heterogeneity of telomerase reverse transcriptase mutation and expression, telomerase activity and telomere length across human cancer cell lines cultured in vitro. Exp. Cell Res. 2020;396:112298. doi: 10.1016/j.yexcr.2020.112298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.