Figure 1.
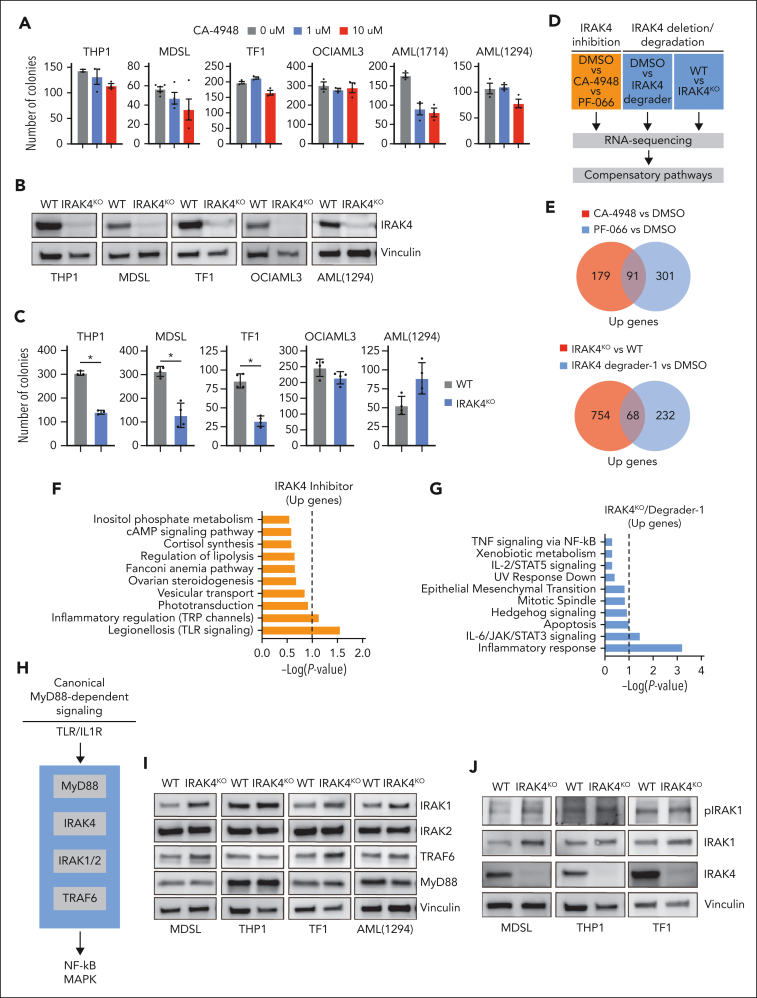
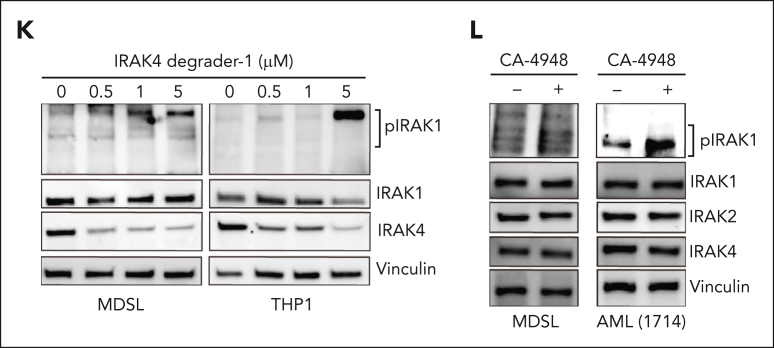
IRAK4 inhibition causes the activation of IRAK1. (A) Colony formation in a panel of MDS/AML cell lines and patient-derived samples treated with the indicated concentrations of CA-4948 (2 independent experiments). (B) Immunoblots for IRAK4 in WT and IRAK4KO AML cell lines and patient-derived samples. (C) Colony formation of WT and IRAK4KO AML cell lines and patient-derived samples. (D) Experimental overview: RNA sequencing was performed using WT and IRAK4KO THP1 cells, and THP1 cells were treated for 24 hours with the indicated inhibitors. Genes upregulated upon IRAK4 deficiency or chemical inhibition were used to annotate compensatory pathways. (E) Venn diagrams of overlapping upregulated genes upon IRAK4 deficiency or IRAK4 chemical inhibition. (F) Pathway enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) data sets using overlapping genes increased upon the treatment with IRAK4 inhibitors. (G) Pathway enrichment of KEGG data sets using overlapping genes increased upon treatment with IRAK4 degrader-1 or after the deletion of IRAK4. (H) Overview of canonical Myd88-dependent signaling: upon TLR ligation, MyD88 nucleates a complex with IRAK4, which signals through IRAK1 and/or IRAK2 and then TRAF6 to activate the NF-κB and MAPK pathways. (I) Immunoblots for IRAK1, IRAK2, TRAF6, and MyD88 in WT and IRAK4KO AML cell lines and patient-derived samples. (J) Immunoblots for phoshpo-IRAK1, total IRAK1, and IRAK4 in WT and IRAK4KO cell lines. (K) Immunoblots for phoshpo-IRAK1, total IRAK1, and IRAK4 in MDSL and THP1 cells treated for 24 hours with IRAK4 degrader-1. (L) Immunoblots for phospho-IRAK1, total IRAK1, IRAK2, and IRAK4 in MDSL and AML (1714) treated for 24 hours with CA-4948 (10 μM). Significance was determined with a Student t test (∗P < .05). Error bars represent the standard deviation.