Figure 6. Overview of the cell envelope chaperone/protease network and of the oxidative protein folding pathway.
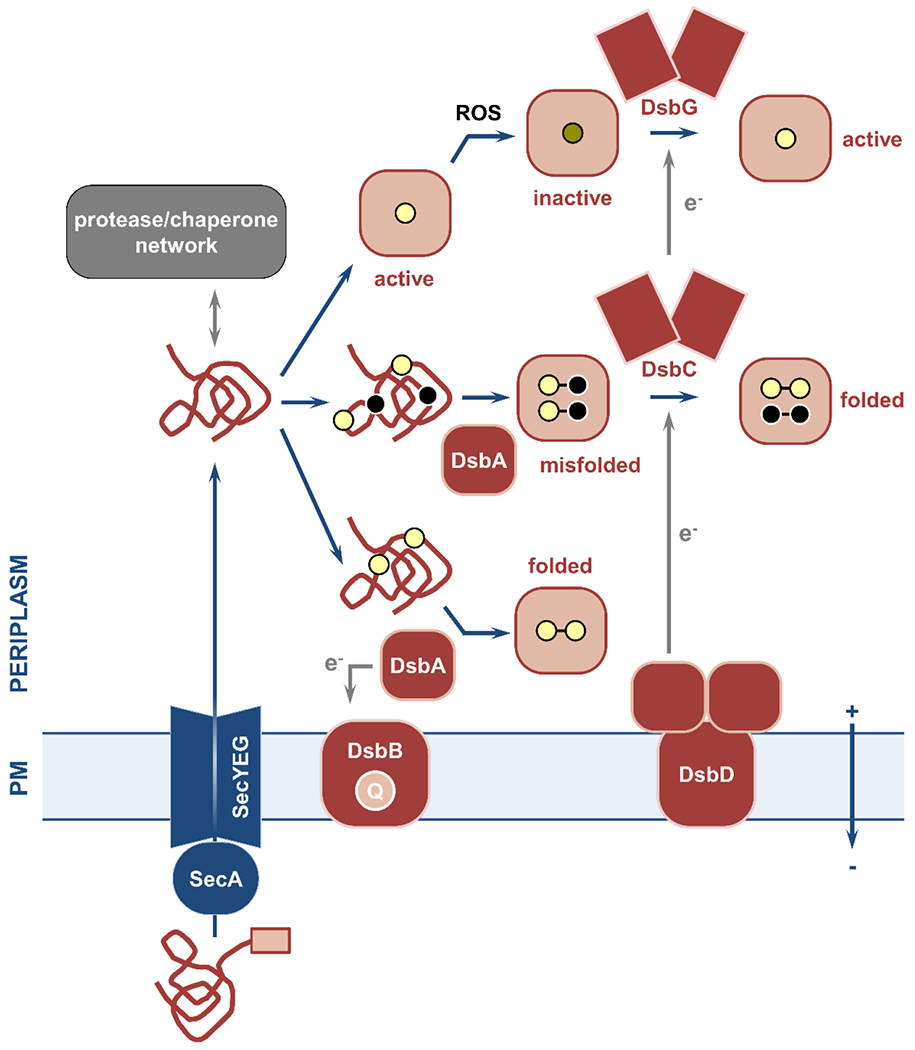
Nascent polypeptides with specific N-terminal signal sequences (pink box) are delivered to the Sec translocon and transported across the plasma membrane. (left) The periplasmic quality control network of chaperones and proteases ensures safe passage of these polypeptides to their final destinations and degradation of misfolded and aggregated proteins, respectively. (right) Oxidative protein folding in Gram-negative bacteria is carried out by the disulfide bond formation system. This process relies on the introduction of disulfide bonds between two consecutive cysteine residues (yellow circles) immediately after Sec-mediated protein translocation. Disulfide formation is catalyzed by the primary oxidase DsbA, which is then reinstated in its active oxidized form by its partner, the quinone (Q)-binding integral membrane protein DsbB. For proteins with only two cysteines (bottom) the introduction of a single disulfide bond results in correct folding. For substrates with more than two cysteines (middle), introduction of bonds in a sequential manner by DsbA may lead to the formation of non-native linkages and protein misfolding. In this case, the isomerase DsbC and its membrane-embedded partner DsbD use cytoplasmic reducing equivalents (e−) to aid refolding of the misfolded protein into its active form. The disulfide bond formation system also includes the reductase DsbG that reverses oxidative damage caused by reactive oxygen species (ROS) to proteins with single cysteines (top). PM, plasma membrane.
