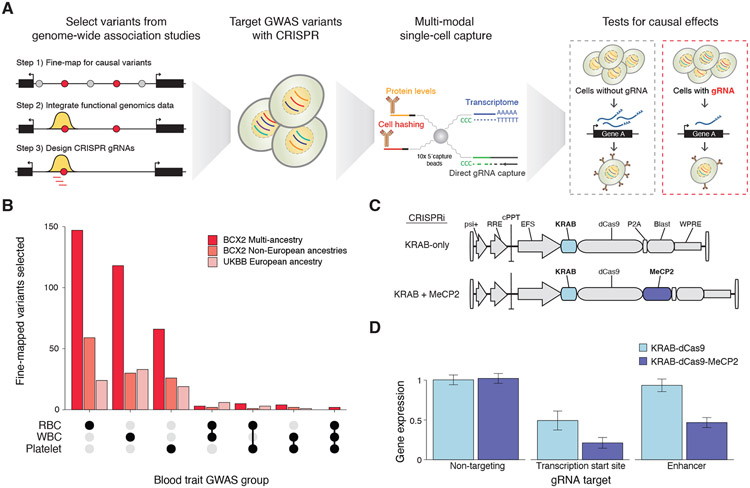
Figure 1. Overview of Systematic Targeting and Inhibition of Noncoding GWAS loci with single-cell sequencing (STING-seq).
(A) STING-seq pipeline for perturbation and single-cell analysis of human genetic variants from genome-wide association studies (GWAS). First, plausibly causal variants are identified via statistical fine-mapping of GWAS. After further refinement of candidate cis-regulatory elements (cCREs) using key molecular hallmarks of regulatory elements, CRISPR guide RNAs (gRNAs) are designed to target cCREs and lentivirally transduced at a high multiplicity-of-infection into human cells. Using multimodal single-cell sequencing, target genes for GWAS variants are identified using differential transcript or protein expression. (B) The number of targeted GWAS variants mapping to cCREs across 29 blood traits in UK Biobank (n = 361,194 participants) and 15 blood traits in the Blood Cell Consortium (n = 746,667 participants). (C) Lentiviral CRISPR inhibition (CRISPRi) vector with a single effector domain (KRAB-dCas9) or dual effector domains (KRAB-dCas9-MeCP2). (D) Mean digital PCR gene expression in human erythrocyte cells (K562) by targeting the transcription start sites (TSS) and known enhancers of three genes (MRPS23, SLC25A27 and FSCN1) with either single-effector KRAB-dCas9 or dual-effector KRAB-dCas9-MeCP2 CRISPRi. Error bars indicate s.e.m.