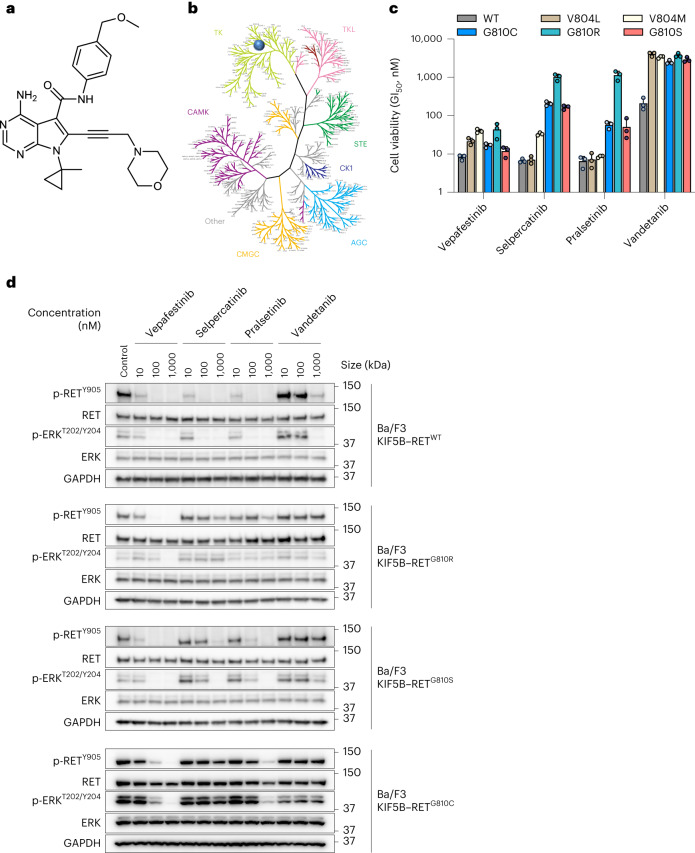
Fig. 1. Structure and biochemical characterization of vepafestinib (TAS0953/HM06).
a, Chemical structure of vepafestinib. b, Kinase selectivity profile of vepafestinib across 255 kinases. Enzyme activities were assessed in the presence of 23 nM vepafestinib, which is approximately 70-fold higher than the IC50 for inhibition of RETWT. Only one kinase (RET) was inhibited by >50% and is shown as a blue circle on the kinome tree. TK, tyrosine kinase; TKL, tyrosine kinase-like; CAMK, calcium/calmodulin-dependent protein kinase; STE, homologs of yeast sterile 7, sterile 11 and sterile 20 kinases; CK1, casein kinase 1; CMGC, cyclin-dependent kinases, mitogen-activated protein kinases, glycogen synthase kinases and cell division control protein-like kinases; AGC, protein kinase A, protein kinase G and protein kinase C families. c, GI50 (50% growth inhibition) values of vepafestinib, in comparison to other RET inhibitors on proliferation of Ba/F3 cells expressing KIF5B–RETWT or KIF5B–RET harboring mutations in the solvent front of the kinase domain (G810R, G810S or G810C) or the gatekeeper domain (V804L or V804M). Data represent the mean ± s.d. of three independent experiments. d, Effect of vepafestinib on phosphorylation of RET and downstream signals in Ba/F3 cells expressing KIF5B–RETWT, KIF5B–RETG810R, KIF5B–RETG810S or KIF5B–RETG810C. Cells expressing KIF5B–RETWT, KIF5B–RETG810R, KIF5B–RETG810S or KIF5B–RETG810C were treated with the indicated concentrations of each drug for 1 h before preparation of cell extracts for western blotting. Representative immunoblots from two independent experiments are shown. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. p, phosphorylated.