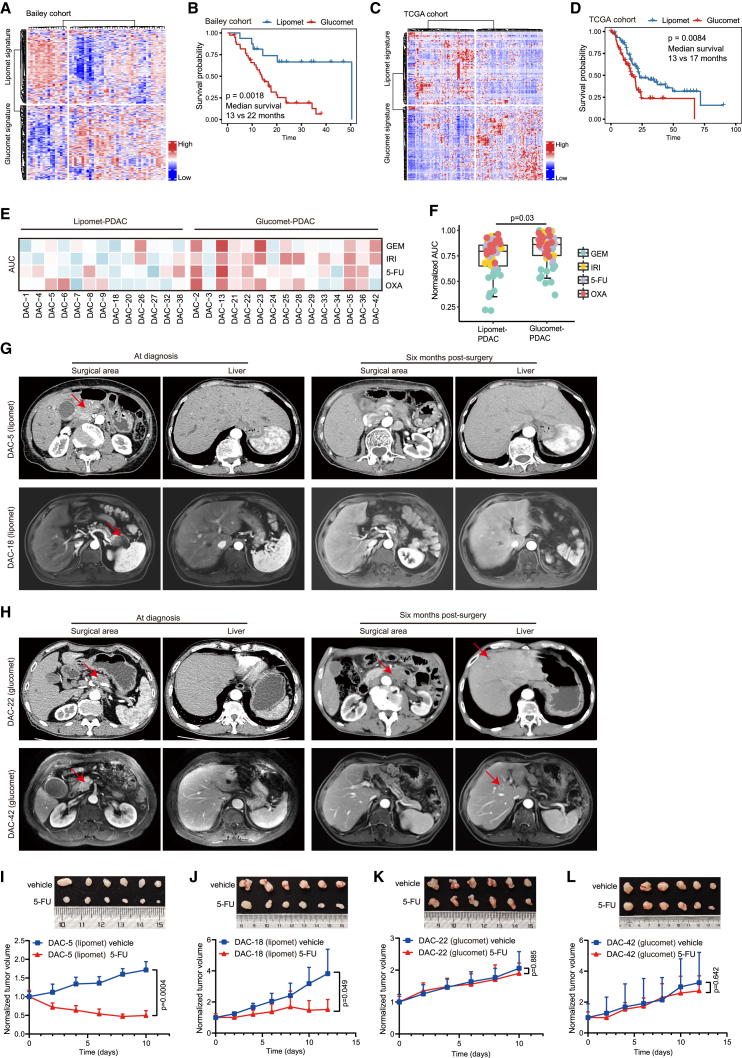
Figure 2.
Glucomet-PDAC is associated with worse prognosis and chemoresistance
(A) Heatmap of tumors in the Bailey PDAC cohort (n = 55, only squamous and pancreatic progenitor samples were included) split by glucomet and lipomet signature genes.
(B) Kaplan-Meier survival curves of the Bailey PDAC cohort showing differential prognosis among patients with different subtypes.
(C) Heatmap of tumors in the TCGA PDAC cohort (n = 156, only ductal pancreatic cancer samples were included) split by glucomet and lipomet signature genes.
(D) Kaplan-Meier survival curves of the TCGA PDAC cohort showing differential prognosis among patients with different subtypes.
(E) Normalized AUC distribution for GEM, 5-FU, OXA, and IRI on glucomet-PDAC (n = 15) and lipomet-PDAC (n = 13). The Z scores of the obtained normalized AUC values are depicted in the heatmap. High values (indicating resistance) are depicted in red, and low values (indicating sensitivity) are depicted in blue.
(F) Comparison of AUCs of four agents among the two metabolic subtypes. The boxplot shows the median (central line) and the 25%–75% interquartile range (box limits).
(G and H) Representative radiation examination of both the surgical area and liver in lipomet-PDAC (DAC-5 and DAC-18) and glucomet-PDAC (DAC-42 and DAC-22) at the time of diagnosis and 6 months postsurgery. The arrow marks the tumor position.
(I–L) 5-FU responsiveness test in the indicated ODX models (n = 6 per group). Tumor volumes measured by calipers at the indicated time points in tumor-implanted mice subjected to treatments with control or 5-FU (25 mg/kg, every 2 days).
Statistical significance was computed by log rank test (B and D). Significance was computed by a one-sided paired t test (F). Data are presented as the mean values ± SEMs, and statistical significance was computed by unpaired Student’s t test (I–L). GEM, gemcitabine; 5-FU, 5-fluorouracil; IRI, irinotecan; OXA, oxaliplatin.