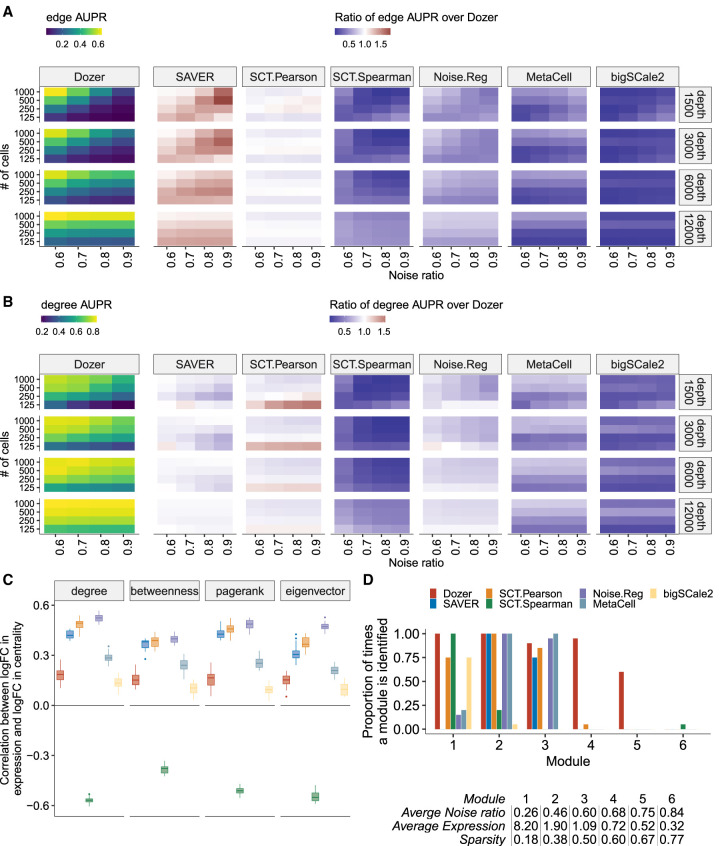
Figure 2.
Evaluation for gene centrality and module estimation. (A,B) Summary of simulation results in terms of AUPR scores for edge and high degree centrality gene identification. The leftmost panel depicts the average AUPR scores of Dozer for edge (A) and degree (B) centrality identification as a function of gene noise ratios (x-axis), number of cells (y-axis), and average sequencing depths (rows). The remaining panels highlight the performances of other methods as quantified by the ratio of their AUPR scores over the AUPR score of Dozer. Results for the identification of high centrality genes with respect to pagerank, betweenness, and eigenvector centrality are in Supplemental Figure S7. (C) Robustness of estimated gene centrality measures against differential expression. Box plots of Spearman's correlations between log2 fold changes (logFCs) of expression and centrality across genes are displayed for individual methods. Although the two batches of simulated data sets have induced differential expression, they share the same coexpression network structure, leading to differential expression but similar centrality measures of the genes across the two batches. (D) Proportion of times each module is identified by each method. A gene module was deemed as identified by a method if it has a Jaccard index overlap of at least 0.5 with WGCNA estimated modules of the method. The table below the bar plot provides the average sparsity, expression, and noise ratio of genes in each module.