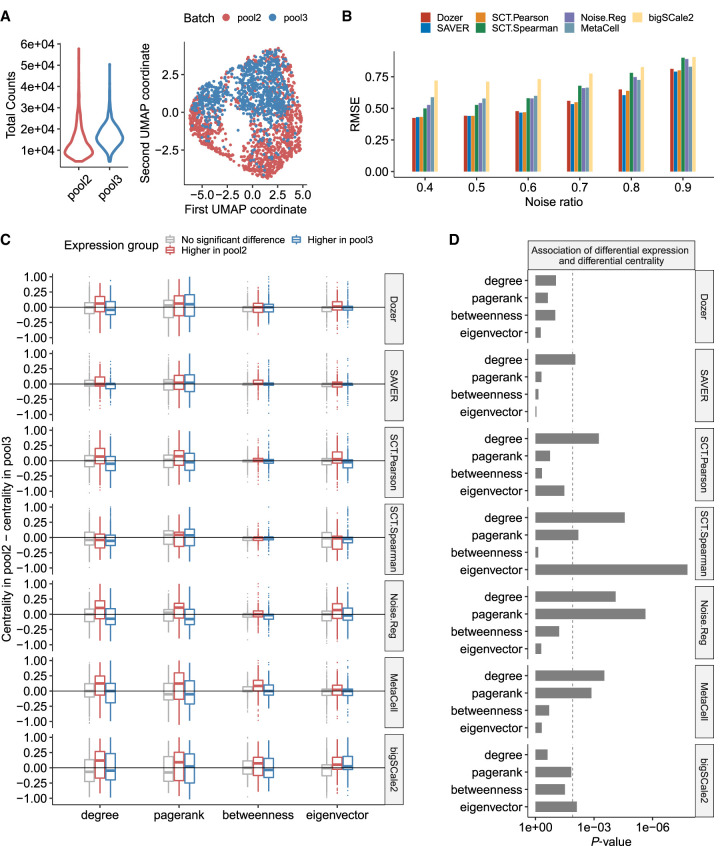
Figure 3.
Robustness in coexpression networks against sequencing depth differences of the scRNA-seq data sets. (A) Distribution of sequencing depths across cells and UMAP visualization of the cells in biological replicates pool2 and pool3. (B) The root mean square error (RMSE) of absolute correlation estimates in pool2 using the higher depth pool3 as the gold standard. Before computing the RMSE, the absolute correlations in pool2 and pool3 were scaled by the standard error of all absolute correlations. (C) Genes are separated into three groups as “higher expression in pool2,” “higher expression in pool3,” and “no significant differential expression” using an adjusted P-value threshold of 0.05. For each gene group, the boxplot displays the differences in gene centrality scores between the pool2 and pool3 data sets. Methods robust to sequencing depth differences have centrality differences centered at zero regardless of the gene group. (D) P-values from testing the association of differential expression and differential centrality. Dashed line is the Bonferroni-corrected P-value threshold of 0.05/4.