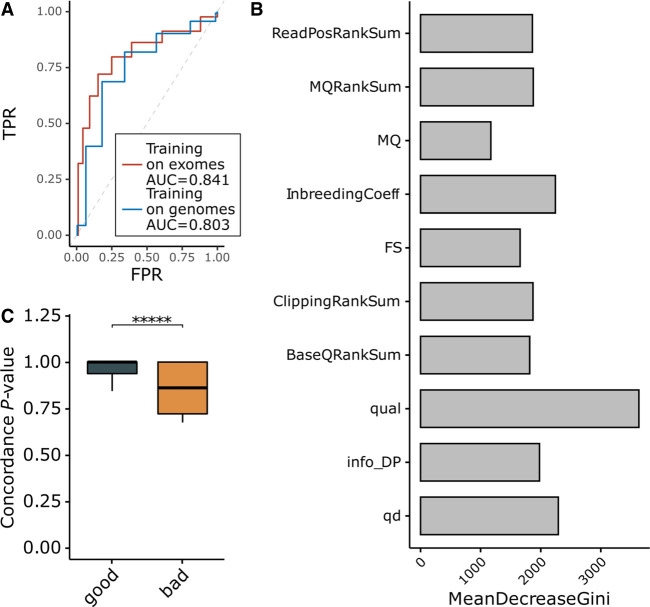
Figure 2.
Reliable classification of discordant variants based on variant annotations. (A) ROC analysis for two random forest predictor validation approaches either using leave-one-out analysis on the exomes or using the genomes as a training set to classify exome variants. (B) Feature importance analysis for the random forest model. (C) Comparison of concordance analysis Fisher's exact test P-values for variants from the 1000 Genomes classified using the random forest predictor trained on the gnomAD genomes data set. MeanDecreaseGini is a measure of how each variable contributes to the homogeneity of the nodes and leaves in the resulting random forest. The higher the value of mean decrease accuracy or mean decrease Gini score, the higher the importance of the variable in the model.