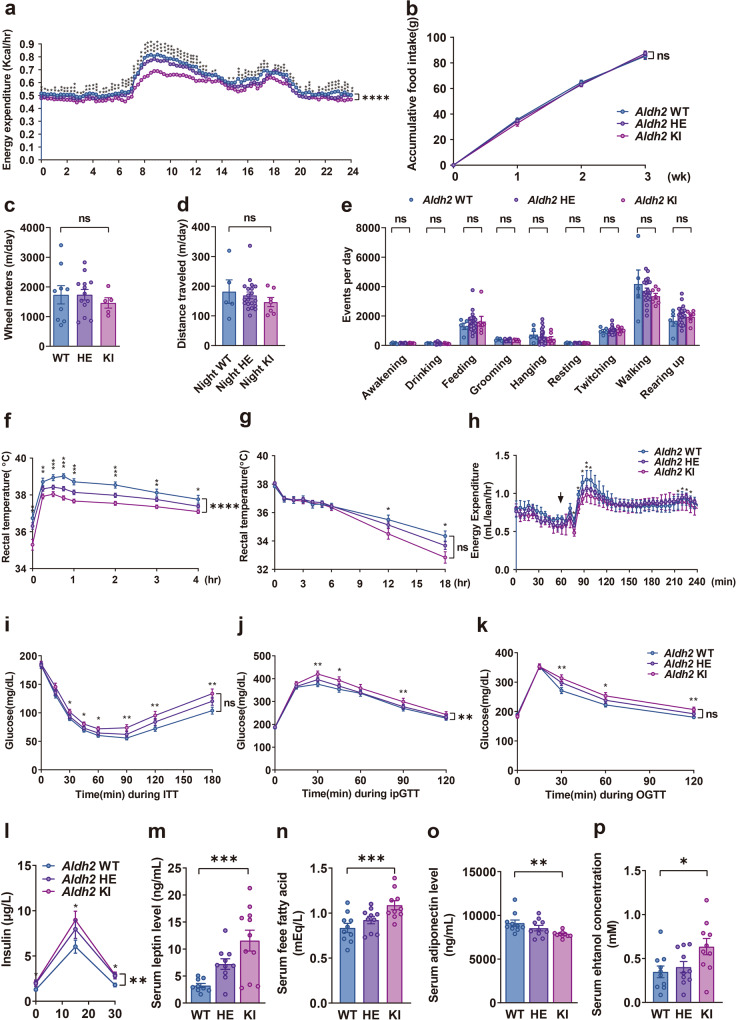
Fig. 2. Aldh2 (acetaldehyde dehydrogenase 2) knock-in mice mimicking human Glu504Lys mutation have reduced energy expenditure and adaptive thermogenesis.
a Energy expenditure (n = 19:27:13; repeated measures analysis of variance [ANOVA] P < 0.0001), b accumulative food intake for three weeks (n = 8:24:10; repeated measures ANOVA P = 0.70), c wheel rotations (n = 9:13:5; P-for-trend = 0.55), d distance traveled (n = 5:20:7 P-for-trend = 0.27), and e behaviors monitored by HomeCage systems (n = 5:20:7; P-for-trend = 0.88, 0.54, 0.48, 0.12, 0.31, 0.92, 0.65, 0.19, and 0.52 respectively) of the Aldh2 homozygous knock-in (KI), heterozygous KI (HE), and wild-type (WT) mice. f Diet-induced thermogenesis measured by rectal temperature after high-fat high-sucrose feeding (repeated measures ANOVA P < 0.0001) and g cold-induced thermogenesis measured by rectal temperature of mice in 4 °C condition (n = 9:24:10; repeated measures ANOVA P = 0.29). h Isoproterenol-induced energy expenditure (n = 4:8:4) of Aldh2 homozygous KI, HE, and WT mice. The arrow head indicates the injection time (repeated measures ANOVA P = 0.15). i Glycemic levels during the insulin sensitivity test (repeated measures ANOVA P = 0.055) and j intraperitoneal glucose tolerance tests (repeated measures ANOVA P = 0.0049), and k glycemic levels during the oral glucose tolerance test of Aldh2 KI, HE, and WT mice (n = 49:24:42; repeated measures ANOVA P = 0.053). l Insulin levels following oral glucose load (n = 35:14:44; repeated measures ANOVA P = 0.0048 in duplicates). Fasting serum m leptin (n = 9:10:11; P-for-trend = 0.0002 in duplicates), n free fatty acid, and (n = 10:10:10; P-for-trend = 0.0008 in duplicates), o adiponectin (n = 10:10:10; P-for-trend = 0.0008 in duplicates), and p ethanol (n = 10:10:10; P-for-trend = 0.0032 in duplicates) concentration of Aldh2 homozygous KI, HE, and WT mice on high-fat high-sucrose diet. Figure (a) was analyzed using the generalized linear model. Figures (a, b, f–l) were further analyzed using repeated measures ANOVA. Figures (b–p) were analyzed using tests for linear trend. All data are presented as mean and standard error (S.E.M.). The n values represent biological repeats and the number of technical repeats were expressed as duplicates or triplicates. The asterisks indicate two-sided *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.001.