Extended Data Fig. 3. Genotyping and characterization of BFD2-deficient parasites.
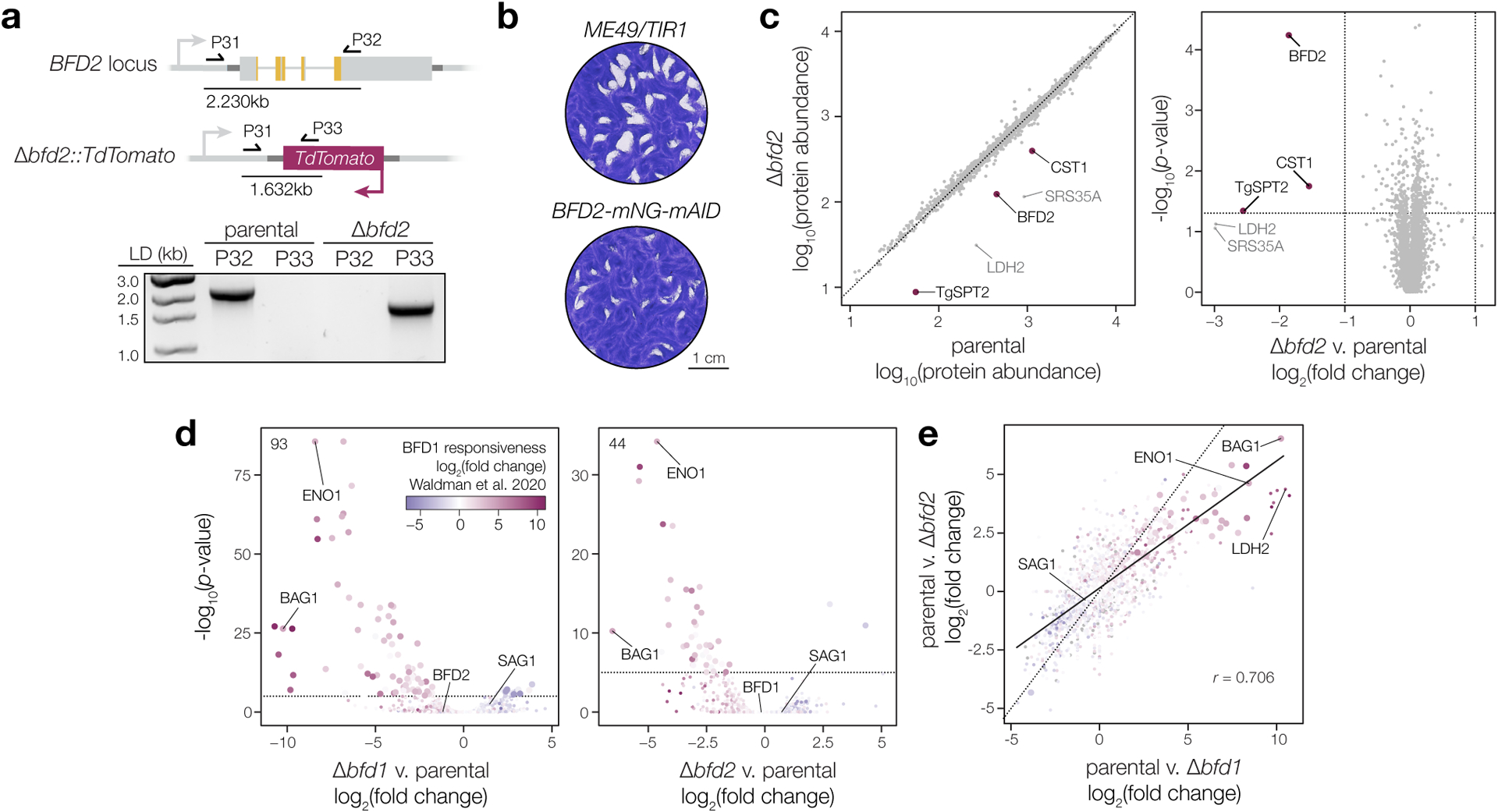
a, Δbfd2 clones were screened by PCR for replacement of the endogenous coding sequence with TdTomato using a common forward primer in the BFD2 5′ UTR (P31) and reverse primers against either BFD2 (P32) or the fluorescent reporter (P33). Diagram shows priming in the parental strain (top) and at the modified allele in Δbfd2 (below), with expected product sizes indicated. Reverse primers and gDNA template used in each reaction are listed above the respective lane. b, Plaque assays after 16 days of undisturbed growth. c, Comparison of protein abundance in unstressed parental (Δbfd1::BFD1-TY) and Δbfd2 parasites. Quantitative proteomics identified a total of 29,806 unique peptides corresponding to 4,303 individual proteins. Significantly affected proteins (magenta) were defined as those meeting three criteria: (i) a minimum of two unique peptides, (ii) absolute fold-change > 2, and (iii) p-value < 0.05. Differences are limited to canonical bradyzoite markers (CST1, LDH2, and SRS35A) or other developmentally regulated genes (TgSPT2)78. d–e, Effects of BFD1 or BFD2 deletion on the parasite transcriptome during infection of mouse primary neurons. Data are based on n = 3 independent infections with color assigned based on log2(fold change) during conditional BFD1 expression23. Significantly affected genes (adjusted p-value < 0.05 by Wald test with DESeq2) are indicated by larger point size. Differential expression analysis was performed for parasites lacking either factor, as compared to the parental strain. Number of genes meeting the cutoff for statistical significance is indicated (d). Comparison of the effects of BFD1- versus BFD2-deletion (e) reveals a comparatively larger impact for the former. Pearson correlation performed on all significant points with trend line fit by linear regression.
