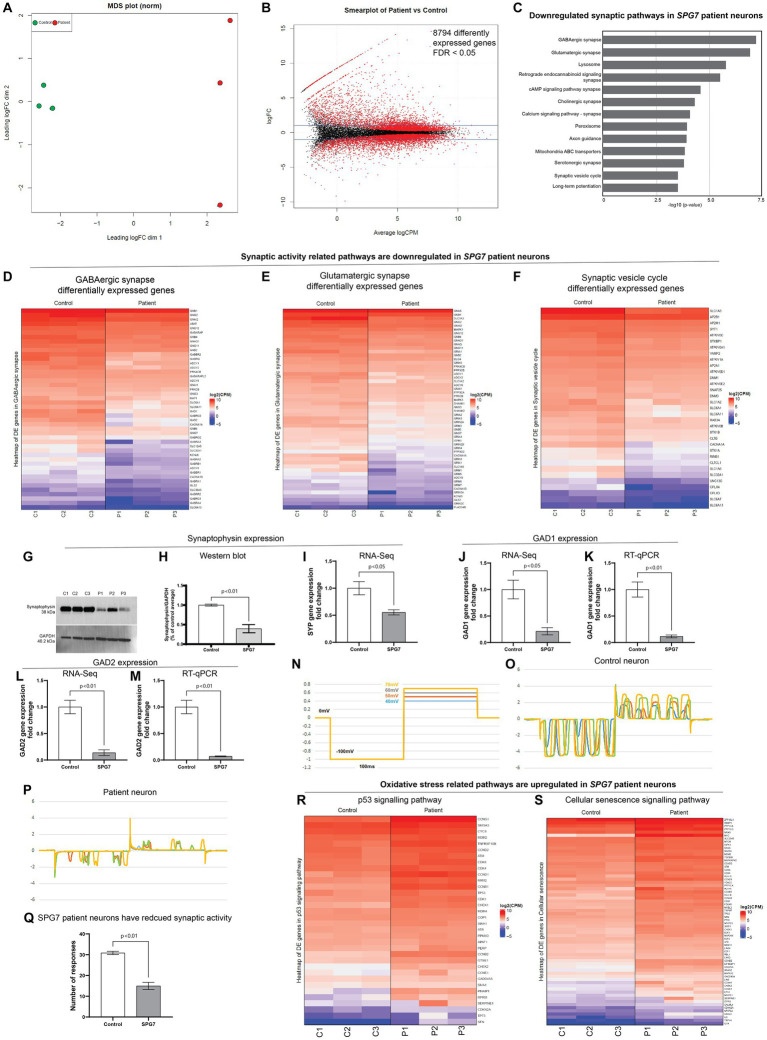
Figure 3.
RNA-Seq analysis of HSP-SPG7 patient neurons. RNA-Seq analysis was performed using patient and healthy control neurons to identify gene expression pathways affected in HSP-SPG7 patient neurons. (A) Multidimensional scaling analysis was used to visualize the level of similarity in the gene expression of the patient and control neurons. (B) Smearplot shows a large number of differently expressed genes. (C) Pathway enrichment analysis was performed to understand which pathways/gene networks the differentially expressed genes are implicated in. Synaptic pathways was FIGURE 3 (Continued)downregulated in patient neurons. (D–F) List of genes that expressed lower in patient neurons compared to control neurons in the GABAergic synapse (D), Glutamergic synapse (E) and synaptic vesicle cycle pathway (F). (G,H) Western blot analysis and (I) RNA-Seq consistently showed reduced expression of synaptophysin expression. (J–M) RT-qPCR validated the findings of RNA-Seq based GAD1 and GAD2 gene expression. (N–Q) Whole cell patch clamping was used to measure neuronal synaptic activity in control and patient neurons. (R,S) List of genes that expressed higher in patient neurons compared to control neurons in the oxidative stress related p53 (R) and cellular senescence (S).