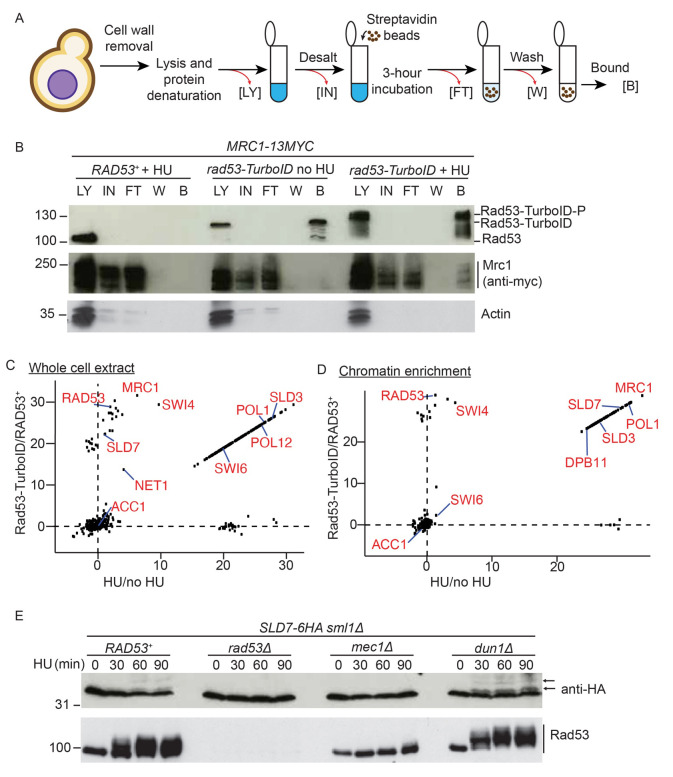
Figure 2. In vivo biotinylation strategy identifies new Rad53-interactors.
A) Protocol for isolation of biotinylated proteins from yeast. LY = lysate, IN = input, FT = flow through, W = wash, B = bound. B) Western blot of the biotinylated purified proteins from the indicated strains, released from G1 phase into either a normal S-phase (no HU) or S-phase in the presence of 200 mM HU (+ HU). C) Label-free quantification (LFQ) analysis of mass spectrometry results from the whole cell extracts. The analysis compares HU treated versus untreated on the x-axis and the Rad53-turboID tagged strain versus the untagged wild type strain on the y-axis. Each dot represents a different statistically significant protein hit. Some key replication factors and known Rad53 interactors/substrates are indicated in red. Acc1, acetyl-CoA carboxylase, is a biotin containing enzyme and an expected hit in all conditions. LFQ values of zero were converted to 1 to allow the calculation of the ratios, which is why many of the hits lie on a diagonal line (see methods). D) As c), except from the chromatin enriched proteome. E) Western blot of HA-tagged Sld7 from the indicated strains. Lower mobility, likely phosphorylated forms of Sld7 are highlighted with arrows.