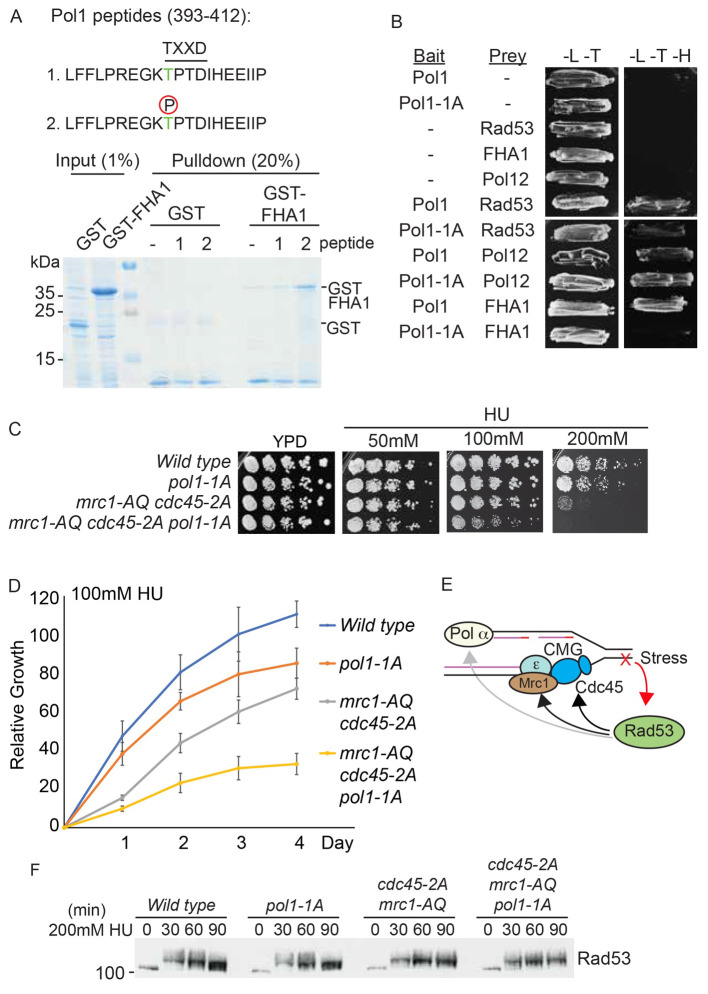
Figure 5. Interaction between phosphorylated residue T402 in Pol1 and Rad53 FHA1 is required for viability in HU.
A) Top, sequence of the Pol1 peptides (393-412) containing the putative FHA binding consensus TXXD, which also contains a CDK consensus site (TP). Peptide 1 is unphosphorylated, peptide 2 contains phospho-threonine at position 402. These peptides contain a N-terminal biotin moiety (not shown). Bottom, Coomassie stain of a streptavidin pulldown of the biotinylated Pol1 peptides incubated with E. coli whole cell extracts expressing either GST or GST fused to the Rad53 FHA1 domain (1-165). B) Yeast two-hybrid analysis of the indicated bait and prey proteins (- indicates empty vector control). All proteins are full length, except for the FHA1 domain of Rad53 (1-165). Pol1-1A contains a T402A mutation at the TXXD consensus site. -L-T media is non-selective, while -L-T-H media is selective for interacting proteins. C) Growth assays of the indicated yeast strains on YPD media, with or without the addition of HU. All plates were grown at 30°C and this image was taken at 48 hours. The mrc1-AQ mutant has all 17 Mec1/Tel consensus phosphorylation sites mutated to alanine and the cdc45-2A mutant has the Rad53 interaction sites (T189/T195) mutated to alanine. D) Quantitation of the 100 mM HU plate growth assay in c) over time. Error bars are SD, n=3. E) Schematic diagram showing interactions of Rad53 with components of the leading strand machinery (Cdc45/Mrc1) and the lagging strand machinery (Pol alpha). Rad53 binds to Cdc45 and Mrc1 and also phosphorylates them (black arrows), while Rad53 binds to, but does not phosphorylate Pol1 (grey arrow). F) Rad53 western blot of the indicated strains released from G1 phase into S-phase with 200mM HU.