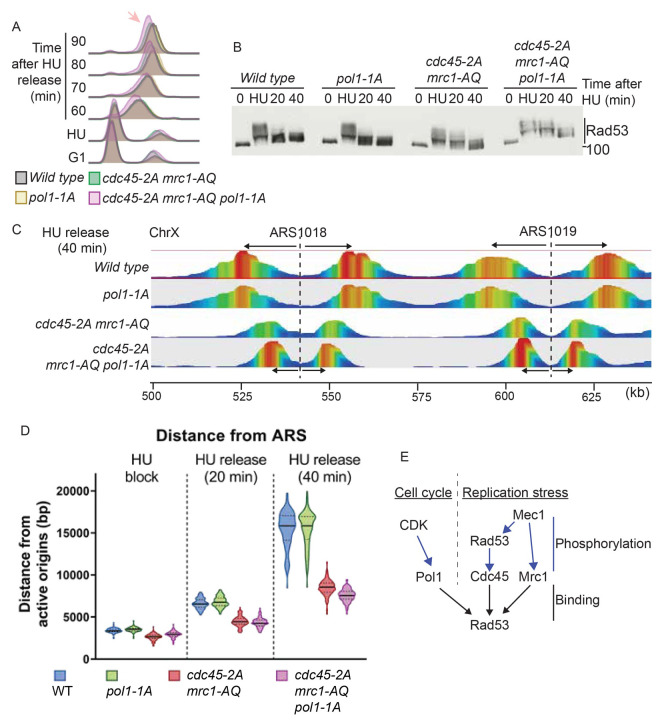
Figure 6. Rad53 interaction with replication proteins, including Pol1, affects fork progression.
A) Flow cytometry of the indicated strains released from G1 phase (0) into S-phase with 200 mM HU for 30 minutes (HU), followed by washing away the HU and release. The arrow indicates a delay in S-phase completion in the cdc45-2A mrc1-AQ pol1-1A strain. B) Rad53 western blot of the experiment in a). C) TrAEL-seq analysis of replication forks 40 minutes after release of the indicated strains from HU arrest. Only a fragment of Chromosome X is shown here. TrAEL-seq reads are coloured by abundance from red to blue. The position of the origins ARS1018 and ARS1019 are indicated with dotted lines and the direction of movement of replication forks from the origins is indicated with arrows. D) Violin plot of the distance of replisomes from origins at the indicated times after release from HU. The average distance travelled by forks from each origin was calculated based on the site of maximum read count within 20 Kb of the origin. The y-axis is to the distance of each replication fork from the corresponding origin. The data here are representative from two biological repeats. The p-values from the averages of these biological replicates is in Supplementary Figure 5. E) Rad53 binds to at least three proteins at the replication fork; Pol1, Cdc45 and Mrc1 (black arrows). The interaction of Rad53 with Mrc1 and Cdc45 is mediated by phosphorylation of these proteins by Mec1 and Rad53 respectively (right, blue arrows). Pol1 on the other hand, is phosphorylated by CDK, which allows binding of Pol1 to Rad53 FHA1 (left). Therefore, the interactions between Rad53 and the replication machinery are regulated both by replication stress (right) and by the cell cycle (left).