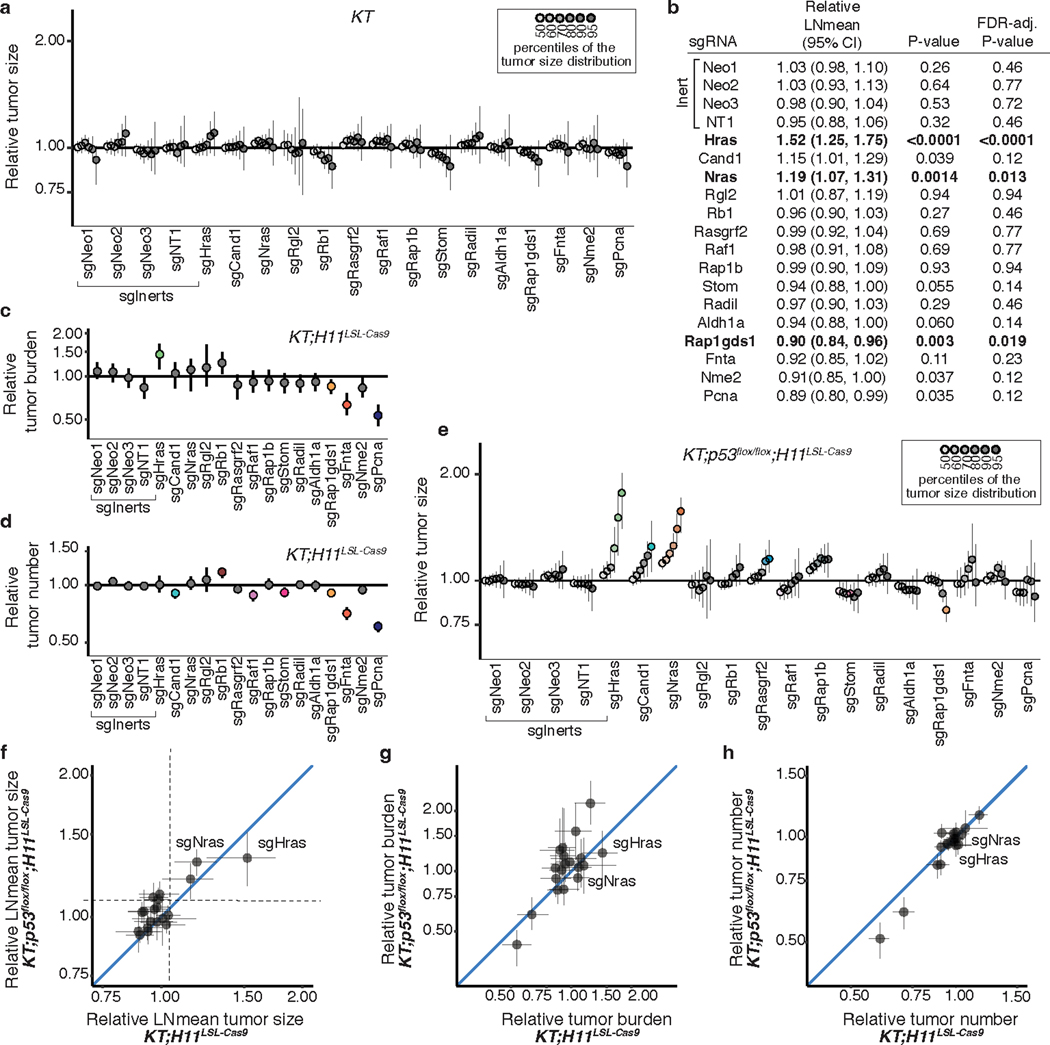
Extended Data Fig. 3. Inactivation of KRAS-interacting proteins has similar impacts on tumor growth in p53-proficient and p53-deficient contexts.
a. Points denote tumor sizes at indicated percentiles for each sgRNA relative to the size of sgInert-containing tumors at the corresponading percentiles in KT mice. Genes are ordered as in Fig. 1f. Line at y = 1 indicates no effect relative to sgInert. As expected, no percentiles were significantly different from sgInert (two-sided FDR-adjusted p < 0.05). b. The impact of each sgRNA on mean tumor size relative to sgInerts in KT;H11LSL-Cas9 mice, assuming a log-normal distribution of tumor sizes (LNmean). sgRNAs with two-sided P < 0.05 after FDR-adjustment are in bold. c-d. Points denote the impact of each sgRNA on tumor burden (c) and number (d) relative to sgInerts in KT;H11LSL-Cas9 mice, normalized to the corresponding statistic in KT mice to account for representation of each sgRNA in the viral pool. sgInerts are in gray and the line at y = 1 indicates no effect. Relative tumor burdens and numbers significantly different from sgInert (two-sided FDR-adjusted p < 0.05) are in color. e. Points denote tumor sizes at the indicated percentiles for each sgRNA relative to the size of sgInert-containing tumors in KT;p53flox/flox;H11LSL-Cas9 mice. Genes are ordered as in Fig. 1f. The line at y = 1 indicates no effect relative to sgInert. Percentiles that are significantly different from sgInert (two-sided FDR-adjusted p < 0.05) are in color. f-h. Comparison of the impact of each sgRNA on relative LNmean tumor size (f), tumor burden (g) and tumor number (h) in KT;H11LSL-Cas9 and KT;p53flox/flox;H11LSL-Cas9 mice. For all panels: Error bars indicate 95% confidence intervals around point estimates of the test statistics. Confidence intervals and P-values were calculated using a nested bootstrap resampling approach described across 11 KT;H11LSL-Cas9 mice, 6 KT;p53flox/flox;H11LSL-Cas9 mice and 5 KT mice.