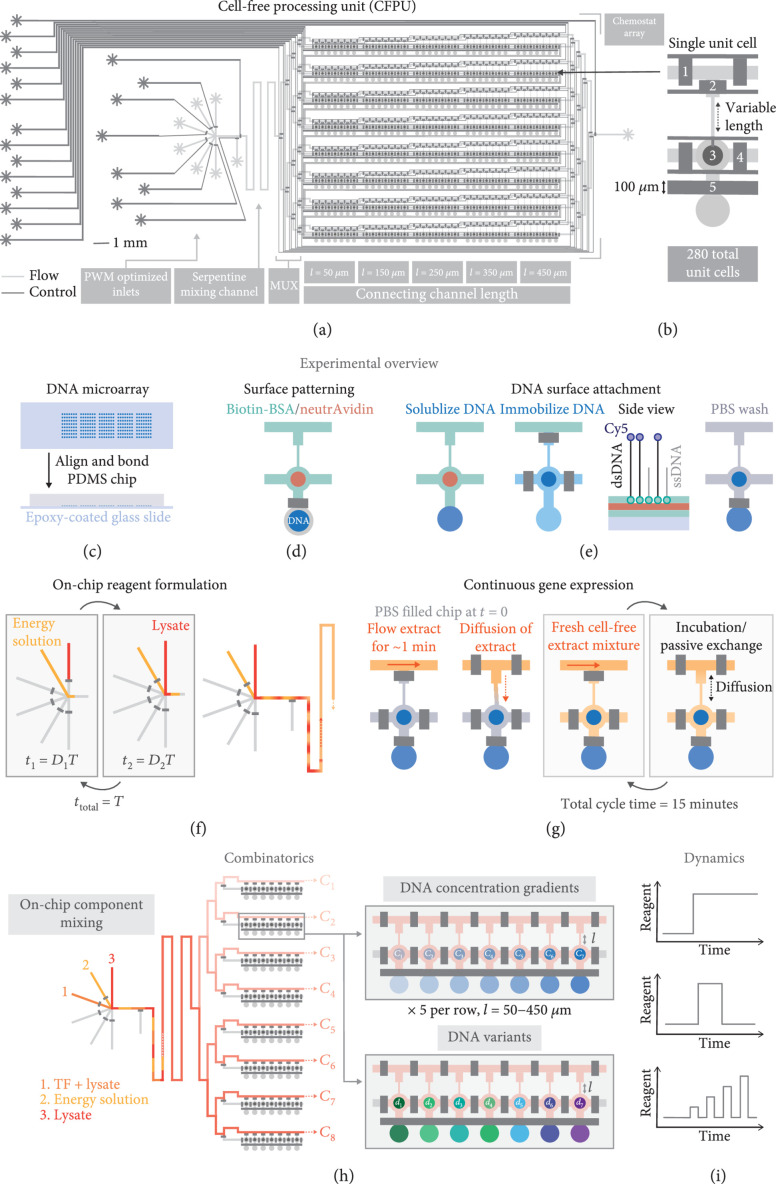
Figure 1.
Overview of the CFPU architecture and functionality. (a) Illustration of the microfluidic CFPU design, highlighting the PWM optimized inlets, followed by the serpentine mixing channel and chemostat array with variable connecting channel lengths. (b) Design of a single unit cell. The pneumatic valves are described from top to bottom: (1) a pair of valves separate each unit cell in the exchange channel, (2) a valve prevents flow into the unit cell while the exchange channel is being replaced with fresh reagents, (3) the button valve enables surface patterning for DNA attachment, (4) sandwich valves separate the reaction chambers of each unit cell from one another, and (5) the neck valve isolates the DNA spot until surface patterning is complete. (c–g) A schematic summary of the experimental protocol: (c) DNA templates are spotted with a microarray robot onto an epoxy-coated glass slide, on top of which the PDMS chip is aligned. (d) The reaction chamber of the unit cell is patterned with biotin-BSA and neutrAvidin. (e) The DNA spot is solubilized, permitting the DNA to diffuse into the upper half of the unit cell and bind to the surface, after which all unbound DNA is washed away. (f) Lysate and energy solutions are then flowed and mixed on-chip with PWM. Each solution is flowed for a time , where and represent the duty cycle and cycle period, respectively. (g) Initially, a cell-free extract solution is flowed onto the chip and diffuses into a unit cell that is filled with PBS. Every 15 minutes, fresh cell-free extract is flowed through the exchange channel for ~1 minute followed by an incubation period that enables the exchange of reagents via diffusion. (h) Examples of combinatoric experiments that can be performed with this chip, including the generation of different concentrations of an input reagent using PWM. Each row of the device can be addressed with a specified reagent concentration, which is then combined with a range of DNA template concentrations or template variants, and different connecting channel lengths. (i) Reagent formulation is dynamic, and a given reagent can be introduced into the device at user-defined times, allowing dynamic perturbations to be performed.