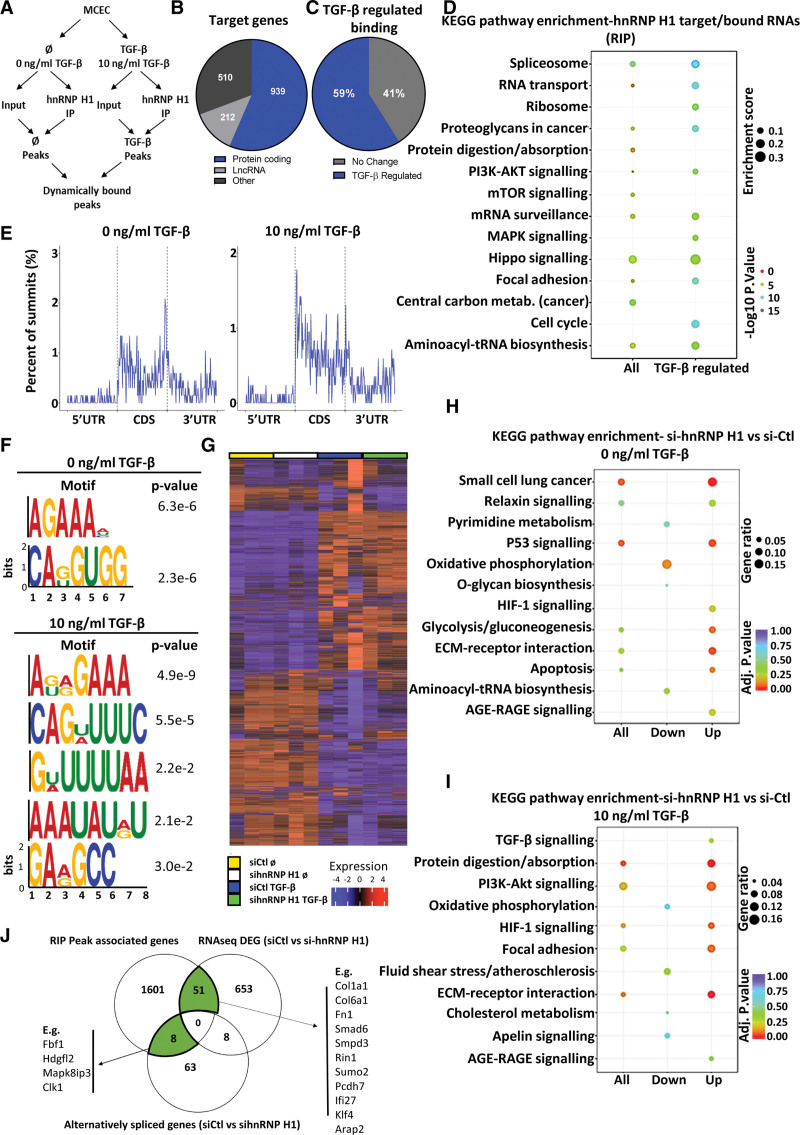
Figure 4.
Identification of the dynamically bound target RNAs of hnRNP H1 (heterogeneous nuclear ribonucleoprotein H1) using RNA immunoprecipitation sequencing (RIP-seq) and RNA-sequencing (RNA-seq). A, Schematic overview of RIP-seq design. Libraries were generated from input and hnRNP H1 immunoprecipitation (IP) samples±TGF (transforming growth factor)-β stimulation to identify global and TGF-β–regulated binding patterns of hnRNP H1. B, Target RNAs of hnRNP H1. Proportion peak-associated genes identified as targets of hnRNP H1, sorted by gene type. C, Proportion of TGF-β–regulated binding. Proportion of hnRNP H1 target genes differentially bound after TGF-β stimulation (P<0.05). D, Pathway enrichment of hnRNP H1 targets. Enriched terms (KEGG pathway) among all peak-related genes (all targets) and differentially bound peak-related genes (TGF-β–regulated binding). E, mRNA summit location. The proportion of summit locations identified within mRNA targets±TGF-β stimulation (from averaged replicates). F, Predicted binding motifs of hnRNP H1. Significantly enriched motifs were identified in peaks in the presence or absence of TGF-β stimulation. G, Heatmap of effects of hnRNP H1 knockdown on RNA expression. RNA-seq after siRNA knockdown (48 hours) of hnRNP H1 in the absence (0 ng/mL TGF-β, siCtl_NS vs sihnRNP_NS) or presence (10 ng/mL TGF-β, siCtl_TGF vs sihnRNP_TGF) of TGF-β stimulation. H, Pathway enrichment of differentially expressed genes si-Ctl vs si-hnRNP H1 in nonstimulated cells. Pathway enrichment among all differentially expressed genes, upregulated and downregulated genes in si-hnRNP H1 samples compared with si-Ctl samples in the absence of TGF-β stimulation. I, Pathway enrichment of differentially expressed genes si-Ctl vs si-hnRNP H1 in TGF-β stimulated cells. Pathway enrichment among all differentially expressed genes, upregulated and downregulated genes in si-hnRNP H1 samples compared with si-Ctl samples in the presence of TGF-β stimulation. K, Overlap between RIP-seq and RNA-seq. The overlap between peak-associated genes identified as targets of hnRNP H1 (RIP-seq) compared with differentially expressed genes (si-Ctl- vs si-hnRNP H1, P<0.05 RNA-seq) and genes with significant alternative splicing (si-Ctl- vs si-hnRNP H1, P<0.05 RNA-seq). AGE-RAGE indicates advanced glycation end-products-receptor for AGE; CDS, coding sequence; DEG, differentially expressed gene; ECM, extracellular matrix; HIF, hypoxia inducible factor; KEGG, Kyoto Encyclopedia of Genes and Genomes; lncRNA, long noncoding RNA; MAPK, mitogen-activated protein kinase; MCEC, mouse cardiac endothelial cell; metab., metabolism; mTOR, mammalian target of rapamycin; siCtl_NS and siCtl_TGF, control siRNA nonstimuated and control siRNA TGF-β stimulated; sihnRNP_NS and sihnRNP_TGF, hnRNP H1 siRNA nonstimulated and hnRNP H1 siRNA TGF-β stimulated; siRNA, small interfering RNA; tRNA, transfer RNA; and UTR, untranslated region.