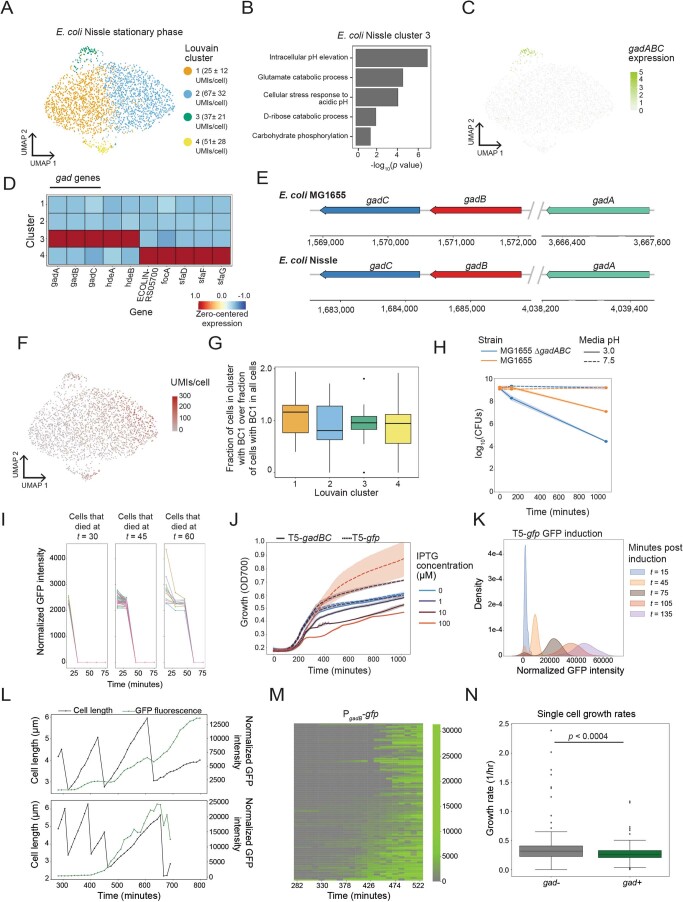
Extended Data Fig. 6. Subpopulation of early stationary phase cells expressing acid-tolerance genes also identified in E. coli Nissle.
a. UMAP of E. coli Nissle transcriptomes from cells at early stationary phase (OD = 2.6). Colours indicate clusters of transcriptionally similar cells. b. GO-term enrichment of select biological processes calculated with marker genes identified for cluster 3 in (A). Marker gene identification and GO term analyses were performed as described in Materials and Methods. c. Same as (A) but with colour gradient indicating expression of gadABC genes (in normalized UMI counts). d. Zero-centred and normalized expression of marker genes for each cluster identified in (A). Marker genes were defined as described in Materials and Methods. e. Schematics of gadABC genes in the two strains of E. coli used in this study: MG1655 and Nissle. f. Same as (A) but with colour gradient indicating number of UMIs captured in each cell. g. Normalized cluster percentage for each BC1 in each cluster (N = 1, 1295, 1053, 83, 68 cells respectively). The normalized percentage for each BC1/cluster combination and boxplot limits are determined as described in Materials and Methods. h. Plot depicts survival of wildtype E. coli MG1655 and ∆gadABC mutant with and without exposure to acid stress during early stationary phase. Curves indicate mean values, and the shaded regions the 95% confidence interval between 2 biological replicates for control samples, and 4 biological replicates for acidified samples. i. Plot depicts fluorescence intensity of individual PgadB-GFP transformed E. coli MG1655 cells during acid exposure as described in Materials and Methods. Fluorescence intensity tracks are broken out by the time of death of each cell. j. Plot depicts growth of E. coli transformed with gadBC (solid) or gfp (dashed) transgene under different concentrations of IPTG inducer. Curves indicate mean values, and the shaded regions the 95% confidence interval between 3 technical replicates for each sample. k. Single-cell fluorescence distributions of E. coli transformed with GFP transgene after induction. l. Representative growth and GFP fluorescence intensity traces of E. coli transformed with PgadB-gfp during growth into stationary phase. m. Fluorescence kymograph of E. coli transformed with PgadB-gfp over time from (K). n. Single-cell growth rates of gad- and gad+ cells from (L,M) using time-lapse microscopy. gad- and gad+ cells were determined as described in Materials and Methods (N = 1, 93, 78 cells respectively). Growth rates were computed as described in Materials and Methods. p = 0.00032 obtained from independent, two-sided t-test.