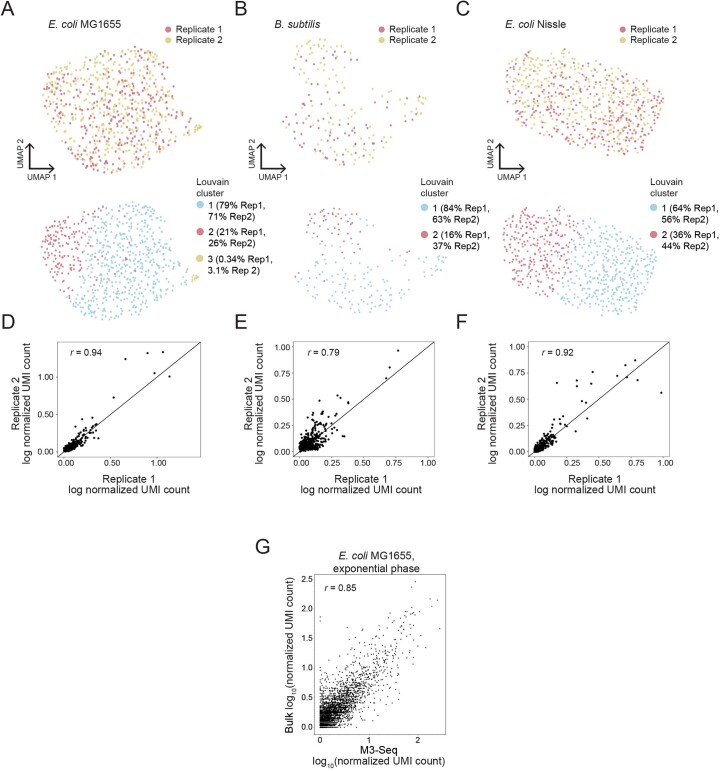
Extended Data Fig. 4. M3-seq profiling during exponential growth and early stationary phase.
a. Top: UMAP of replicate M3-seq data generated from E. coli MG1655 treated with twice the minimum inhibitory concentration of ciprofloxacin, sampled after 6 hours of treatment. b. Same as (A) but for B. subtilis 168. c. Same as (A) but for E. coli Nissle. d. Comparison of replicate data from (A) using mean log normalized UMI counts per cell (that is, unique UMIs relative to total UMIs per cell averaged across all cells for each gene). Each point represents a single gene. r, Pearson correlation. e. Same as (D) but using data from (B). f. Same as (D) but using data from (C). g. Comparison of RNA-seq data to M3-seq pseudobulk profiles from exponential phase E. coli from eBW3. Pseudobulk measurements were obtained by normalizing UMI counts by the total number of UMIs in the dataset and log transforming the normalized counts. Each point represents a single gene. r, Pearson correlation.