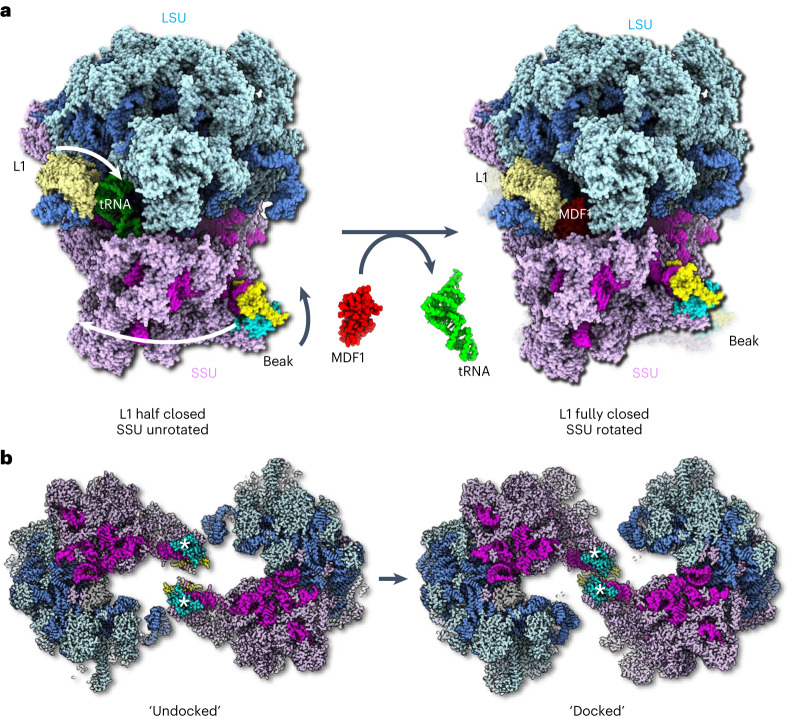
Fig. 5. Conformational differences between the tRNA-bound 70S ribosome and the MDF1-bound hibernating 100S ribosome.
a, Left: the structure of the (uninhibited) 70S ribosome in atom representation. The L1 stalk (blue, cream) is bound to an E-site tRNA (lime green) and is in a ‘half-closed'’ conformation. The small ribosomal subunit (SSU, magenta and pink) is in an unrotated state. The beak is in a straightened position. Right: atomic model of a half-dimer within a hibernating 100S particle. The E-site tRNA is replaced with the putative MDF1 protein (red). The L1 stalk is in a ‘fully-closed’ conformation and the SSU is in a rotated state. The beak is bent towards the large ribosomal subunit (LSU, shades of blue). Arrows in the left panel indicate conformational changes of L1, SSU and beak as the ribosome transitions from tRNA bound to MDF1 bound. b, Left: the tRNA-bound structure superimposed with the subtomogram averaging map of the 100S dimer shows a gap between the beaks (*). The beaks are in an ‘undocked’ conformation. Right: atomic model of the 100S ribosome dimer. Rotation of the SSU and bending of the beaks establishes close contact between the beaks, which are now in a 'docked' conformation. An animated version of this figure can be found in Supplementary Video 1.