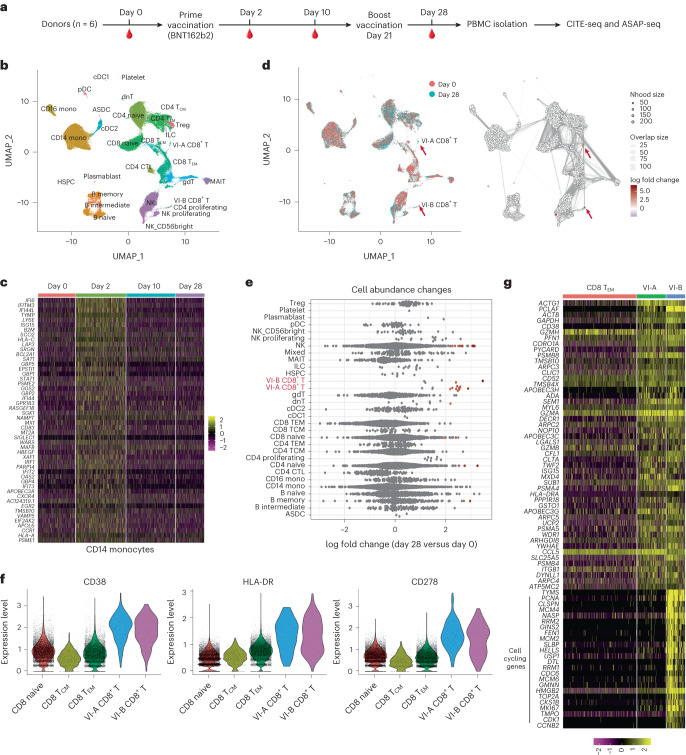
Fig. 1. Multimodal identification of SARS-CoV-2 mRNA vaccine-induced CD8+ T cells.
a, Overview of study design, in which PBMCs were collected from six healthy donors at day 0, day 2, day 10 and day 28 during the BNT162b2 mRNA vaccination process and comprehensive analyses using both CITE-seq and ASAP-seq were performed on these samples. b, Uniform manifold approximation and projection (UMAP) visualizations of 113,897 single cells obtained as in a profiled with CITE-seq and clustered by a weighted combination of RNA and protein modalities. Cells are colored based on level-2 annotation (level-1 and level-3 annotations; Extended Data Fig. 2a). c, Single-cell heatmap showing activation of interferon response module within CD14+ monocytes. d, Milo analysis of differentially abundant cell states between day 0 and day 28 samples as in panel a. UMAP on the left is color coded by timepoint. Right plot indicates embedding of the Milo differential abundance. Each node represents a neighborhood, node size is proportional to the number of cells, and neighborhoods are colored by the level of differential abundance. Nhood, neighborhood. e, Beeswarm plot showing the log-fold distribution of cell abundance changes between day 0 and day 28 samples as in a. Neighborhoods overlapping the same cell population are grouped together, neighborhoods exhibiting differential abundance are colored in red. f, Violin plots with protein upregulation of CD38, HLA-DR and CD278 (ICOS) in VI-A and VI-B CD8+ T cells compared with CD8+ naive, CD8+ TCM and CD8+ TEM cells. g, Heatmap showing mRNA expression of 50 marker genes for VI-A CD8+ T cells, as well as cell-cycling genes highly expressed in VI-B CD8+ T cells. For visualization purposes, a randomly selected subset of CD8+ TEM cells are presented. HSPC, hematopoietic stem and progenitor cell; NK, natural killer; pDC, plasma dendritic cell; TCM, central memory T cell; TEM, effector memory T cell. ASDC, AXL+ dendritic cell; CTL, cytotoxic T cell; dnT, double-negative T cell; gdT, gamma-delta T cell; MAIT, mucosal associated invariant T.