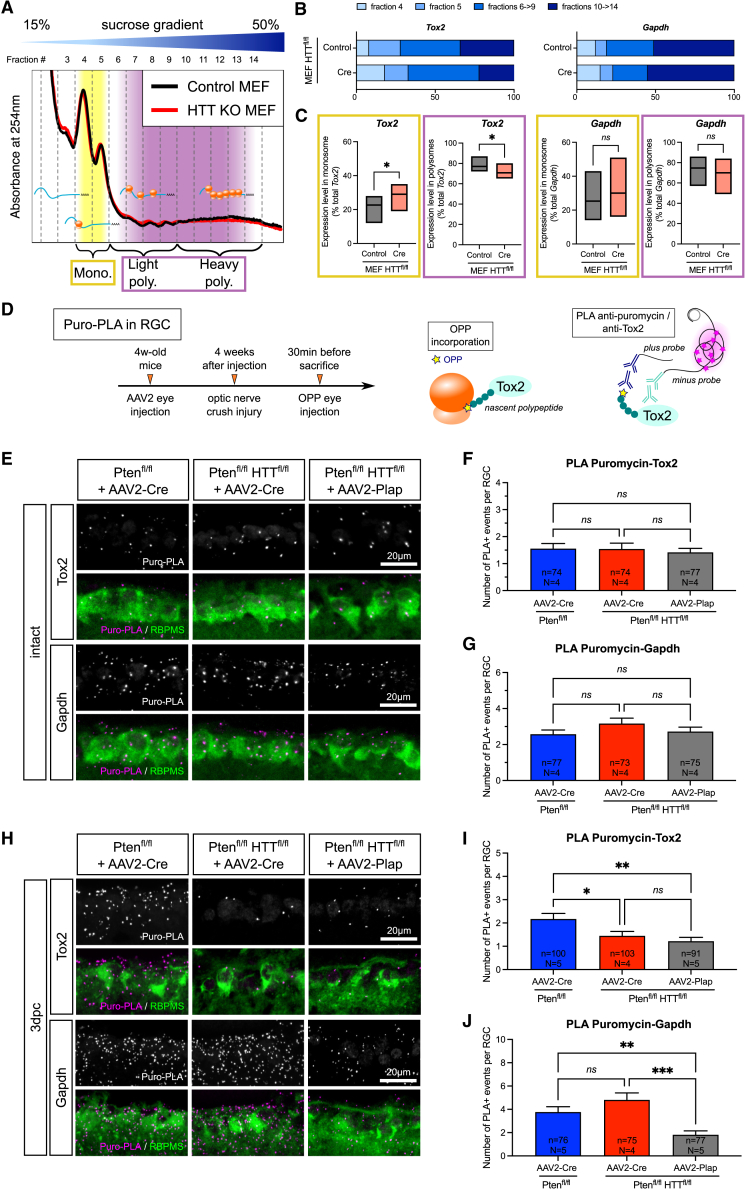
Figure 6.
HTT controls axon regeneration via the translational regulation of Tox2 mRNA
(A) Representative polysome profile of HTT-deleted (KO) MEF versus control on a 15%–50% sucrose gradient.
(B) Representative Tox2 and Gapdh mRNA distribution in fractions 4–14 of a polysome fractionation, in HTT-deleted (Cre) versus control MEFs.
(C) Tox2 and Gapdh mRNA distribution in monosomal (yellow) and polysomal (purple) fractions. Data are represented as min/max values with line at mean. Paired t test; ∗p value < 0.05; ns, not significant.
(D) Principle of puro-PLA experiment in vivo.
(E) PLA staining confocal images for puromycin-Tox2 or puromycin-Gapdh in intact RGCs in Ptenfl/fl+AAV2-Cre, Ptenfl/flHTTfl/fl+AAV2-Cre, and Ptenfl/flHTTfl/fl+AAV2-Plap.
(F) Quantification of puromycin-Tox2 PLA-positive events in Ptenfl/fl+AAV2-Cre, Ptenfl/flHTTfl/fl+AAV2-Cre, and Ptenfl/flHTTfl/fl+AAV2-Plap intact RGCs.
(G) Quantification of puromycin-Gapdh PLA-positive events in Ptenfl/fl+AAV2-Cre, Ptenfl/flHTTfl/fl+AAV2-Cre, and Ptenfl/flHTTfl/fl+AAV2-Plap intact RGCs.
(H) PLA staining confocal images for puromycin-Tox2 or puromycin-Gapdh in Ptenfl/fl+AAV2-Cre, Ptenfl/flHTTfl/fl+AAV2-Cre, and Ptenfl/flHTTfl/fl+AAV2-Plap RGCs at 3dpc.
(I) Quantification of puromycin-Tox2 PLA-positive events in Ptenfl/fl+AAV2-Cre, Ptenfl/flHTTfl/fl+AAV2-Cre, and Ptenfl/flHTTfl/fl+AAV2-Plap RGCs at 3dpc.
(J) Quantification of puromycin-Gapdh PLA-positive events in Ptenfl/fl+AAV2-Cre, Ptenfl/flHTTfl/fl+AAV2-Cre, and Ptenfl/flHTTfl/fl+AAV2-Plap RGCs at 3dpc.
Data are represented as mean ± SEM. Kruskal-Wallis with Dunn’s multiple comparisons test; ∗p value < 0.05, ∗∗p value < 0.01, and ∗∗∗p value < 0.001; ns: not significant.