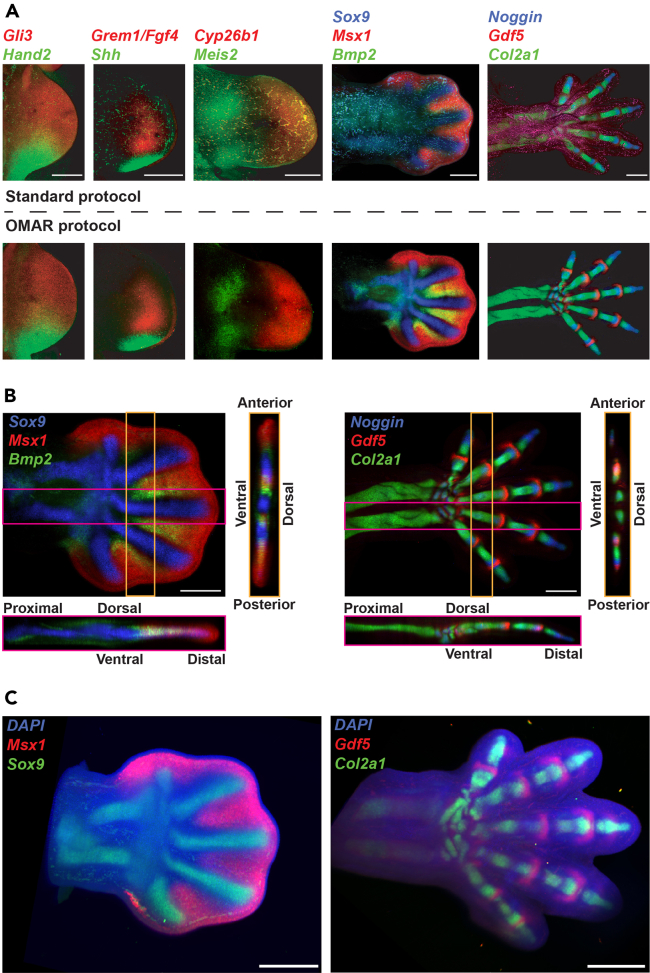
Figure 2.
High signal-to-noise ratio and sensitive detection of multiple spatially distinct gene expression patterns in limb buds of different developmental stages
(A–C) All limb buds shown in (A and B) are processed for and imaged by confocal microscopy without subsequent digital autofluorescence subtraction or masking. (A) Blood cell and blood vessel autofluorescence and probe penetration problems increase progressively with the more advanced stages of mouse limb bud development using a standard HCR RNA-FISH protocol (upper panels). In contrast, OMAR photobleaching in combination with SDS-detergent permeabilization reduces tissue autofluorescence improves probe penetration and signal-to-noise ratio at all stages analyzed (lower panels). (B) 3D reconstruction of the limb buds shown in the two rightmost images (lower panels in A) reveals the tissue integrity, even hybridization/signal detection, and very high signal-to-noise ratio throughout the limb buds. Scale bars: 300 μm. (C) Limb buds first analyzed by confocal microscopy (panels A and C) were remounted and imaged by light-sheet microscopy. Left panel: 3D volume rendering of a forelimb bud to detect both Msx1 (red channel) and Sox9 (green channel). Right panel: 3D volume rendering of a forelimb bud to detect Gdf5 (red) and Col2a1 (green). Nuclei are counterstained by DAPI (blue channel). Methods video S1 and S2 are available online for panel C. Scale bars: 500 μm.