Fig. 6.
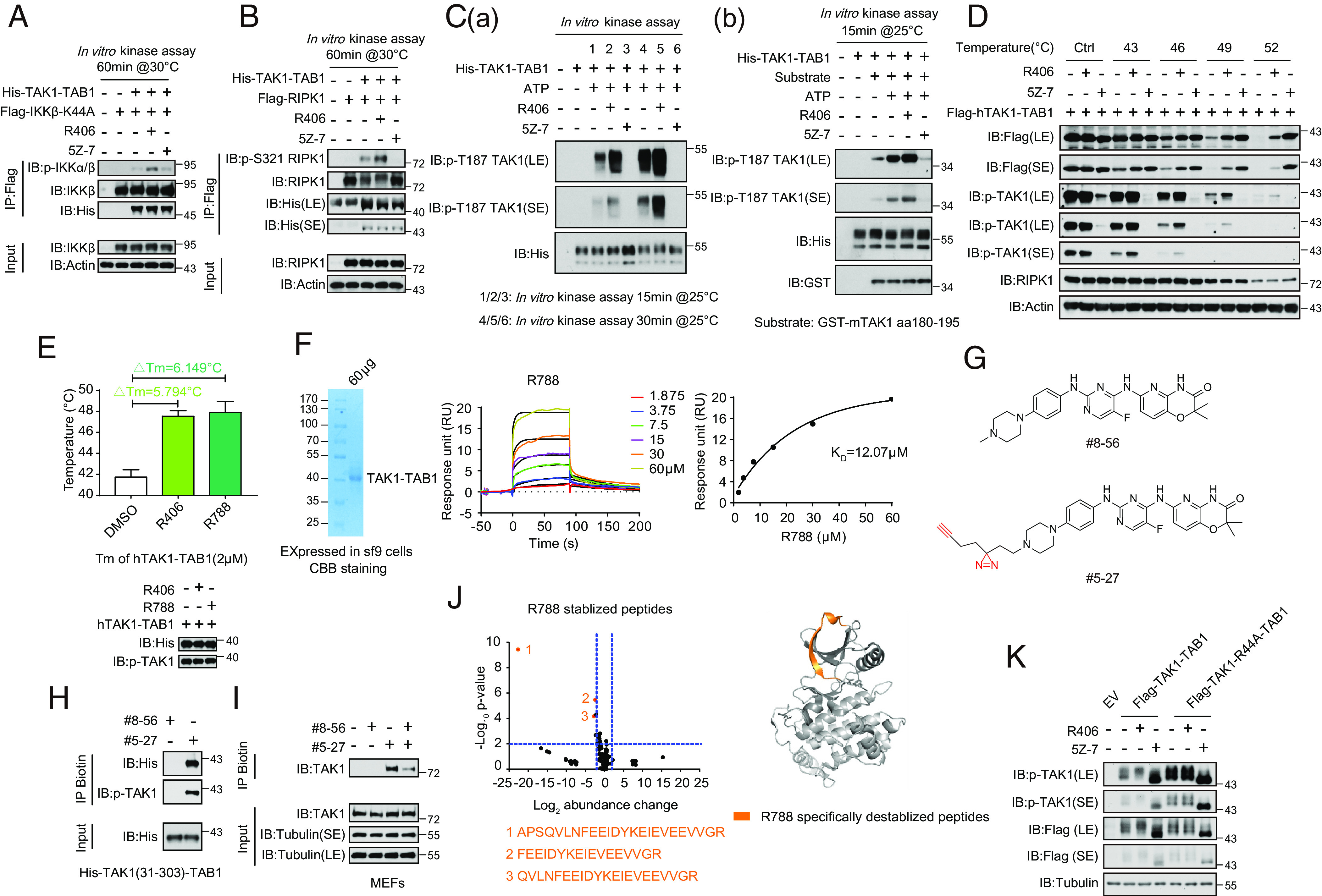
R406/R788 directly interacts with TAK1. (A) Flag-IKKβ (K44A) was expressed and purified from HEK293T cells and incubated with recombinant TAK1-TAB1 (2 μM) or R406 (20 μM), 5Z-7 (1 μM) as control. The products were analyzed by western blotting using indicated antibodies. (B) Flag-RIPK1 was expressed and purified from HEK293T cells and incubated with recombinant TAK1-TAB1 (2 μM) or R406 (20 μM), 5Z-7 (1 μM) as control. The products were analyzed by western blotting using indicated antibodies. (C, a) R406 promotes TAK1 autophosphorylation at Thr187. The TAK1 kinase activity was measured by incubating His-TAK1-TAB1 protein (2 μM) with R406 (20 μM) or 5Z-7 (1 μM) at 25 °C for 15 min and 30 min in the presence of ATP, respectively. The samples were analyzed by immunoblotting with p-TAK1 (Thr187). (b) The TAK1 kinase activity was measured by incubating His-TAK1-TAB1 protein with 4 μg GST fused mouse TAK1 peptide (aa180-195) in vitro in the presence of ATP or R406 or 5Z-7 at 25 °C for 15 min. The samples were analyzed by immunoblotting with p-TAK1(Thr187). (D). Cellular thermal stability. HEK293T cells were transfected with expression vectors for Flag-tagged hTAK1(15-303)-TAB1 fusion expression plasmids for 24 h. 3μM R406 and 500 nM 5Z-7 were added at 20 h after transfection. The HEK293T cells were harvested and resuspended with PBS and then were incubated in 0, 43, 46, 49, 52 °C for 3 min, and then frozen in liquid nitrogen quickly. The cells were subject to repeated freeze–thaw three times, and then centrifuged at 15000rpm for 15min. The products were analyzed by western blot using indicated antibodies. (E) Thermal stability profiles of protein thermal shift assay. The recombinant hTAK1(15-303)-TAB1 fusion protein (2 μM) were purified from the Bac-to-BacTM Baculovirus Expression System and treated with 160 μM R406, R788 for 2 h. The protein thermal stability was analyzed using differential scanning calorimetry by real-time PCR, and the melting temperatures were calculated by Protein Thermal Shift Software. Four replicates for each reaction were performed. (F) Kinetic profile of binding of R788 to TAK1-TAB1 from SPR analysis. (G) The chemical structures of the photoaffinity probe compound #5-27 for R406 and the photostable control compound #8-56. (H) Purified TAK1-TAB1(IIe31-Gln303, His468-Pro504) were incubated with #8-56 (200 μM) or #5-27 (200 μM) for 60 min. Samples were subjected to photo-cross-linking assay and enriched by streptavidin-coupled beads. All isolated complexes were analyzed by western blotting with indicated antibodies. (I) MEFs were pretreated with #8-56 (20 μM) or #5-27 (20 μM) or #8-56 with #5-27 for 1 h, and the cells were lysed with Nonidet P-40 buffer. Samples were subjected to photo-cross-linking assay and enriched by streptavidin-coupled beads. All immunoprecipitated complexes and whole-cell lysates were analyzed by western blotting with indicated antibodies. (J) Volcano plot showing peptides identified by LiP-MS. Recombinant hTAK1(15-303)-TAB1(468-504) purified from Sf-9 cells was treated with vehicle, R788 (200 μM), respectively, for 2 h at room temperature and then subjected to LiP-MS analysis. Abundance change of peptides in R788 treatment group compared to control group was presented as volcano plots. The x axis shows Log2 fold changes and the y axis shows −Log10 P values. Destabilized LiP-peptides were cherry picked in R788 treatment group with P-value<0.05 and abundance change>2 and further categorized into different subgroups. R788 destabilized peptides were colored orange (Left). R788 destabilized peptides were mapped to aligned structure of R788 with TAK1-TAB1 in right. R788 specifically destabilized peptides were colored orange. (K) HEK293T cells were transfected with expression vectors for Flag-tagged TAK1-TAB1 and Flag-TAK1-R44A-TAB1 fusion expression plasmid for 24 h, respectively. 3 μM R406 and 500 nM 5Z-7 were added at 20 h after transfection. The HEK293T cells were lysed with RIPA buffer and analyzed by western blotting with indicated antibodies.
